
Niminivirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; unclassified Geminiviridae
Average proteome isoelectric point is 8.51
Get precalculated fractions of proteins
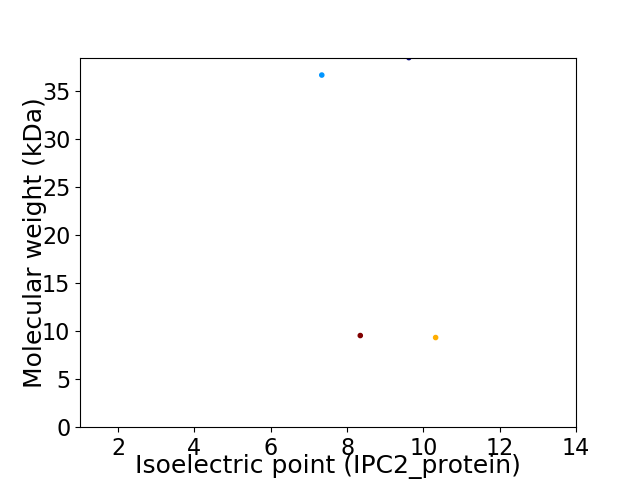
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|J7LLL3|J7LLL3_9GEMI Replication-associated protein OS=Niminivirus OX=1229325 PE=3 SV=2
MM1 pKa = 7.61QYY3 pKa = 10.93PNMPFRR9 pKa = 11.84IQGRR13 pKa = 11.84HH14 pKa = 5.61LFLTYY19 pKa = 8.88PQCHH23 pKa = 6.3LTHH26 pKa = 6.18EE27 pKa = 5.15AIYY30 pKa = 10.79NHH32 pKa = 7.29LIDD35 pKa = 5.48LPVGNEE41 pKa = 3.84KK42 pKa = 8.85PTKK45 pKa = 9.73IVVAKK50 pKa = 9.61EE51 pKa = 3.73LHH53 pKa = 6.52EE54 pKa = 5.35DD55 pKa = 3.71GHH57 pKa = 5.91PHH59 pKa = 5.29FHH61 pKa = 7.77AYY63 pKa = 10.46LCFDD67 pKa = 3.22KK68 pKa = 11.0RR69 pKa = 11.84RR70 pKa = 11.84DD71 pKa = 3.43LRR73 pKa = 11.84YY74 pKa = 9.45QRR76 pKa = 11.84HH77 pKa = 5.36FDD79 pKa = 2.97ISGYY83 pKa = 9.98HH84 pKa = 6.59PNIQVCRR91 pKa = 11.84SVKK94 pKa = 10.44DD95 pKa = 3.3VLKK98 pKa = 10.94YY99 pKa = 7.9VTKK102 pKa = 10.71DD103 pKa = 3.15GEE105 pKa = 4.39YY106 pKa = 9.17MSNFTVIKK114 pKa = 8.46PTIHH118 pKa = 6.1QLIEE122 pKa = 3.98RR123 pKa = 11.84ADD125 pKa = 4.1DD126 pKa = 3.6EE127 pKa = 4.9TSFALSALEE136 pKa = 5.2HH137 pKa = 6.16YY138 pKa = 9.82DD139 pKa = 4.37KK140 pKa = 11.69NFANCFNSWMSLYY153 pKa = 11.13KK154 pKa = 9.74MLKK157 pKa = 9.62SKK159 pKa = 10.84KK160 pKa = 9.39KK161 pKa = 9.39VCDD164 pKa = 3.79PVSDD168 pKa = 3.66WTLMKK173 pKa = 9.97MDD175 pKa = 5.56DD176 pKa = 3.73LHH178 pKa = 8.26LLARR182 pKa = 11.84ILSITAHH189 pKa = 5.84QKK191 pKa = 10.41DD192 pKa = 3.62GSRR195 pKa = 11.84TKK197 pKa = 10.66SIWLFGPSKK206 pKa = 9.69TGKK209 pKa = 8.34STLARR214 pKa = 11.84SIGKK218 pKa = 8.19HH219 pKa = 6.03AYY221 pKa = 7.54MQNTWCVDD229 pKa = 3.54EE230 pKa = 5.79LSDD233 pKa = 4.36DD234 pKa = 3.71AQYY237 pKa = 11.73LVLDD241 pKa = 5.28DD242 pKa = 4.93IAWDD246 pKa = 3.52TWKK249 pKa = 9.36WQYY252 pKa = 11.53KK253 pKa = 9.89SVLGCQKK260 pKa = 10.94DD261 pKa = 3.76VIFTGKK267 pKa = 9.04YY268 pKa = 8.43ARR270 pKa = 11.84PRR272 pKa = 11.84RR273 pKa = 11.84FNFNMPCIVCSNEE286 pKa = 3.74KK287 pKa = 10.2PVFSVDD293 pKa = 3.05EE294 pKa = 4.46LNWLDD299 pKa = 3.57VNIDD303 pKa = 3.52FFEE306 pKa = 4.77INNKK310 pKa = 9.95LYY312 pKa = 10.99
MM1 pKa = 7.61QYY3 pKa = 10.93PNMPFRR9 pKa = 11.84IQGRR13 pKa = 11.84HH14 pKa = 5.61LFLTYY19 pKa = 8.88PQCHH23 pKa = 6.3LTHH26 pKa = 6.18EE27 pKa = 5.15AIYY30 pKa = 10.79NHH32 pKa = 7.29LIDD35 pKa = 5.48LPVGNEE41 pKa = 3.84KK42 pKa = 8.85PTKK45 pKa = 9.73IVVAKK50 pKa = 9.61EE51 pKa = 3.73LHH53 pKa = 6.52EE54 pKa = 5.35DD55 pKa = 3.71GHH57 pKa = 5.91PHH59 pKa = 5.29FHH61 pKa = 7.77AYY63 pKa = 10.46LCFDD67 pKa = 3.22KK68 pKa = 11.0RR69 pKa = 11.84RR70 pKa = 11.84DD71 pKa = 3.43LRR73 pKa = 11.84YY74 pKa = 9.45QRR76 pKa = 11.84HH77 pKa = 5.36FDD79 pKa = 2.97ISGYY83 pKa = 9.98HH84 pKa = 6.59PNIQVCRR91 pKa = 11.84SVKK94 pKa = 10.44DD95 pKa = 3.3VLKK98 pKa = 10.94YY99 pKa = 7.9VTKK102 pKa = 10.71DD103 pKa = 3.15GEE105 pKa = 4.39YY106 pKa = 9.17MSNFTVIKK114 pKa = 8.46PTIHH118 pKa = 6.1QLIEE122 pKa = 3.98RR123 pKa = 11.84ADD125 pKa = 4.1DD126 pKa = 3.6EE127 pKa = 4.9TSFALSALEE136 pKa = 5.2HH137 pKa = 6.16YY138 pKa = 9.82DD139 pKa = 4.37KK140 pKa = 11.69NFANCFNSWMSLYY153 pKa = 11.13KK154 pKa = 9.74MLKK157 pKa = 9.62SKK159 pKa = 10.84KK160 pKa = 9.39KK161 pKa = 9.39VCDD164 pKa = 3.79PVSDD168 pKa = 3.66WTLMKK173 pKa = 9.97MDD175 pKa = 5.56DD176 pKa = 3.73LHH178 pKa = 8.26LLARR182 pKa = 11.84ILSITAHH189 pKa = 5.84QKK191 pKa = 10.41DD192 pKa = 3.62GSRR195 pKa = 11.84TKK197 pKa = 10.66SIWLFGPSKK206 pKa = 9.69TGKK209 pKa = 8.34STLARR214 pKa = 11.84SIGKK218 pKa = 8.19HH219 pKa = 6.03AYY221 pKa = 7.54MQNTWCVDD229 pKa = 3.54EE230 pKa = 5.79LSDD233 pKa = 4.36DD234 pKa = 3.71AQYY237 pKa = 11.73LVLDD241 pKa = 5.28DD242 pKa = 4.93IAWDD246 pKa = 3.52TWKK249 pKa = 9.36WQYY252 pKa = 11.53KK253 pKa = 9.89SVLGCQKK260 pKa = 10.94DD261 pKa = 3.76VIFTGKK267 pKa = 9.04YY268 pKa = 8.43ARR270 pKa = 11.84PRR272 pKa = 11.84RR273 pKa = 11.84FNFNMPCIVCSNEE286 pKa = 3.74KK287 pKa = 10.2PVFSVDD293 pKa = 3.05EE294 pKa = 4.46LNWLDD299 pKa = 3.57VNIDD303 pKa = 3.52FFEE306 pKa = 4.77INNKK310 pKa = 9.95LYY312 pKa = 10.99
Molecular weight: 36.7 kDa
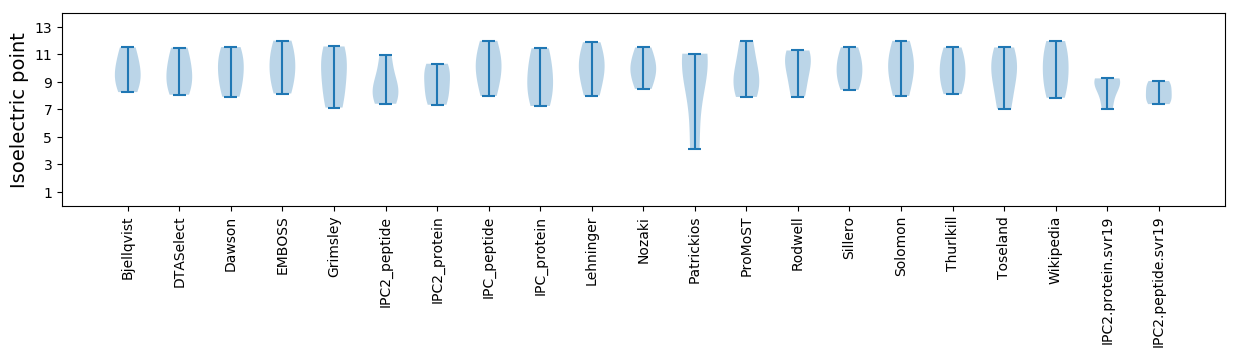
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7LJE9|J7LJE9_9GEMI Putative movement protein OS=Niminivirus OX=1229325 PE=4 SV=2
MM1 pKa = 7.44KK2 pKa = 10.07RR3 pKa = 11.84PSFFSVAKK11 pKa = 10.56AGFKK15 pKa = 10.4RR16 pKa = 11.84YY17 pKa = 10.04GPTIARR23 pKa = 11.84KK24 pKa = 9.2VGKK27 pKa = 9.22HH28 pKa = 4.5ALKK31 pKa = 10.44YY32 pKa = 10.24GYY34 pKa = 10.14RR35 pKa = 11.84KK36 pKa = 8.46LTQSGKK42 pKa = 7.16STRR45 pKa = 11.84EE46 pKa = 4.03TAPLTYY52 pKa = 10.94DD53 pKa = 3.27NDD55 pKa = 3.33FKK57 pKa = 11.06TDD59 pKa = 3.34YY60 pKa = 9.97RR61 pKa = 11.84YY62 pKa = 11.02KK63 pKa = 10.74RR64 pKa = 11.84MPKK67 pKa = 9.22RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84WKK74 pKa = 10.39RR75 pKa = 11.84FTKK78 pKa = 10.23KK79 pKa = 9.87VNAVINRR86 pKa = 11.84SQGLKK91 pKa = 9.86KK92 pKa = 10.47VLYY95 pKa = 9.87SDD97 pKa = 4.03IVRR100 pKa = 11.84LSSNVSQCAYY110 pKa = 9.74HH111 pKa = 6.81SAMLYY116 pKa = 9.77TPDD119 pKa = 4.71ARR121 pKa = 11.84VTDD124 pKa = 4.53LSADD128 pKa = 3.16LGMIFRR134 pKa = 11.84DD135 pKa = 3.14ILGAVAYY142 pKa = 10.78NDD144 pKa = 3.3IQNILVQGDD153 pKa = 3.46VDD155 pKa = 3.66KK156 pKa = 11.19KK157 pKa = 10.95IRR159 pKa = 11.84FEE161 pKa = 4.12SANMDD166 pKa = 3.53VSWRR170 pKa = 11.84NVGANPVIIDD180 pKa = 4.15LYY182 pKa = 9.79YY183 pKa = 9.54VRR185 pKa = 11.84CRR187 pKa = 11.84KK188 pKa = 8.09TFGLTGADD196 pKa = 3.86ADD198 pKa = 4.58NNTQGIFSLGFVKK211 pKa = 10.51QGVIVDD217 pKa = 4.11EE218 pKa = 4.48EE219 pKa = 4.55DD220 pKa = 3.75GNTVGSNRR228 pKa = 11.84QFALTVGSTPFQSSLFCQTYY248 pKa = 10.47RR249 pKa = 11.84ILSKK253 pKa = 10.73KK254 pKa = 10.1RR255 pKa = 11.84ITIAPGNTVSMTLKK269 pKa = 10.49DD270 pKa = 3.68SRR272 pKa = 11.84NKK274 pKa = 9.6VVNAMDD280 pKa = 3.35TRR282 pKa = 11.84ARR284 pKa = 11.84ICMRR288 pKa = 11.84NLTHH292 pKa = 7.35GYY294 pKa = 9.98LFQLYY299 pKa = 9.0GVPGLNGDD307 pKa = 4.01VPVTALASDD316 pKa = 3.98VVFSVQKK323 pKa = 10.47RR324 pKa = 11.84YY325 pKa = 10.18GFYY328 pKa = 10.9LPVSGKK334 pKa = 9.96DD335 pKa = 3.15QTSNIPLL342 pKa = 3.76
MM1 pKa = 7.44KK2 pKa = 10.07RR3 pKa = 11.84PSFFSVAKK11 pKa = 10.56AGFKK15 pKa = 10.4RR16 pKa = 11.84YY17 pKa = 10.04GPTIARR23 pKa = 11.84KK24 pKa = 9.2VGKK27 pKa = 9.22HH28 pKa = 4.5ALKK31 pKa = 10.44YY32 pKa = 10.24GYY34 pKa = 10.14RR35 pKa = 11.84KK36 pKa = 8.46LTQSGKK42 pKa = 7.16STRR45 pKa = 11.84EE46 pKa = 4.03TAPLTYY52 pKa = 10.94DD53 pKa = 3.27NDD55 pKa = 3.33FKK57 pKa = 11.06TDD59 pKa = 3.34YY60 pKa = 9.97RR61 pKa = 11.84YY62 pKa = 11.02KK63 pKa = 10.74RR64 pKa = 11.84MPKK67 pKa = 9.22RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84WKK74 pKa = 10.39RR75 pKa = 11.84FTKK78 pKa = 10.23KK79 pKa = 9.87VNAVINRR86 pKa = 11.84SQGLKK91 pKa = 9.86KK92 pKa = 10.47VLYY95 pKa = 9.87SDD97 pKa = 4.03IVRR100 pKa = 11.84LSSNVSQCAYY110 pKa = 9.74HH111 pKa = 6.81SAMLYY116 pKa = 9.77TPDD119 pKa = 4.71ARR121 pKa = 11.84VTDD124 pKa = 4.53LSADD128 pKa = 3.16LGMIFRR134 pKa = 11.84DD135 pKa = 3.14ILGAVAYY142 pKa = 10.78NDD144 pKa = 3.3IQNILVQGDD153 pKa = 3.46VDD155 pKa = 3.66KK156 pKa = 11.19KK157 pKa = 10.95IRR159 pKa = 11.84FEE161 pKa = 4.12SANMDD166 pKa = 3.53VSWRR170 pKa = 11.84NVGANPVIIDD180 pKa = 4.15LYY182 pKa = 9.79YY183 pKa = 9.54VRR185 pKa = 11.84CRR187 pKa = 11.84KK188 pKa = 8.09TFGLTGADD196 pKa = 3.86ADD198 pKa = 4.58NNTQGIFSLGFVKK211 pKa = 10.51QGVIVDD217 pKa = 4.11EE218 pKa = 4.48EE219 pKa = 4.55DD220 pKa = 3.75GNTVGSNRR228 pKa = 11.84QFALTVGSTPFQSSLFCQTYY248 pKa = 10.47RR249 pKa = 11.84ILSKK253 pKa = 10.73KK254 pKa = 10.1RR255 pKa = 11.84ITIAPGNTVSMTLKK269 pKa = 10.49DD270 pKa = 3.68SRR272 pKa = 11.84NKK274 pKa = 9.6VVNAMDD280 pKa = 3.35TRR282 pKa = 11.84ARR284 pKa = 11.84ICMRR288 pKa = 11.84NLTHH292 pKa = 7.35GYY294 pKa = 9.98LFQLYY299 pKa = 9.0GVPGLNGDD307 pKa = 4.01VPVTALASDD316 pKa = 3.98VVFSVQKK323 pKa = 10.47RR324 pKa = 11.84YY325 pKa = 10.18GFYY328 pKa = 10.9LPVSGKK334 pKa = 9.96DD335 pKa = 3.15QTSNIPLL342 pKa = 3.76
Molecular weight: 38.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
821 |
78 |
342 |
205.3 |
23.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.725 ± 0.573 | 1.949 ± 0.504 |
6.456 ± 0.825 | 2.436 ± 0.91 |
5.725 ± 1.696 | 5.116 ± 1.169 |
2.801 ± 1.099 | 5.359 ± 0.257 |
7.065 ± 0.872 | 9.013 ± 1.014 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.558 ± 0.261 | 4.872 ± 0.484 |
4.263 ± 0.494 | 3.41 ± 0.32 |
7.186 ± 1.314 | 8.283 ± 1.467 |
5.847 ± 0.86 | 6.456 ± 1.319 |
1.218 ± 0.605 | 4.263 ± 0.675 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
