
Lake Sarah-associated circular virus-4
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
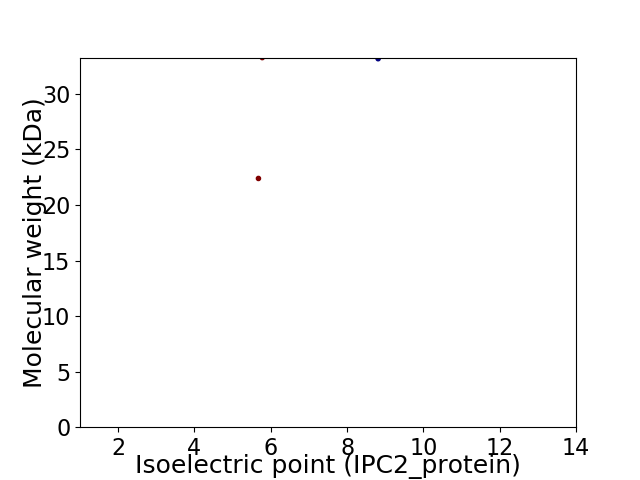
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA19|A0A126GA19_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-4 OX=1685768 PE=4 SV=1
MM1 pKa = 6.8TQRR4 pKa = 11.84ATCWSITINNPTDD17 pKa = 3.39EE18 pKa = 4.42EE19 pKa = 4.8TKK21 pKa = 10.57VALPVGWKK29 pKa = 8.48LTGQIEE35 pKa = 4.19QGEE38 pKa = 4.86EE39 pKa = 4.39GTVHH43 pKa = 5.06YY44 pKa = 10.62QGMLTTTQQRR54 pKa = 11.84FGSIKK59 pKa = 9.6VYY61 pKa = 10.16FPRR64 pKa = 11.84AHH66 pKa = 6.73IEE68 pKa = 3.66VARR71 pKa = 11.84NKK73 pKa = 10.04KK74 pKa = 9.75ALSEE78 pKa = 4.16YY79 pKa = 8.28VHH81 pKa = 7.22KK82 pKa = 10.95SDD84 pKa = 3.81TRR86 pKa = 11.84VAKK89 pKa = 10.7VDD91 pKa = 4.09DD92 pKa = 5.09NISNNIFQWQAEE104 pKa = 4.45VAKK107 pKa = 10.16LWNDD111 pKa = 2.98TDD113 pKa = 4.21FTHH116 pKa = 6.49MKK118 pKa = 10.09YY119 pKa = 10.12IYY121 pKa = 10.15DD122 pKa = 4.07KK123 pKa = 11.08EE124 pKa = 4.16PSEE127 pKa = 4.84DD128 pKa = 3.45VALKK132 pKa = 10.82YY133 pKa = 10.61VDD135 pKa = 4.0SLCGKK140 pKa = 9.91LIRR143 pKa = 11.84EE144 pKa = 4.29GAKK147 pKa = 10.0GLEE150 pKa = 4.21FVAVNPMWRR159 pKa = 11.84SSWKK163 pKa = 9.8RR164 pKa = 11.84FYY166 pKa = 10.99FSIITRR172 pKa = 11.84DD173 pKa = 3.51GRR175 pKa = 11.84DD176 pKa = 3.11EE177 pKa = 4.29AVCEE181 pKa = 4.18TQVPQGEE188 pKa = 4.38EE189 pKa = 4.08TNEE192 pKa = 4.01KK193 pKa = 10.74GSS195 pKa = 3.46
MM1 pKa = 6.8TQRR4 pKa = 11.84ATCWSITINNPTDD17 pKa = 3.39EE18 pKa = 4.42EE19 pKa = 4.8TKK21 pKa = 10.57VALPVGWKK29 pKa = 8.48LTGQIEE35 pKa = 4.19QGEE38 pKa = 4.86EE39 pKa = 4.39GTVHH43 pKa = 5.06YY44 pKa = 10.62QGMLTTTQQRR54 pKa = 11.84FGSIKK59 pKa = 9.6VYY61 pKa = 10.16FPRR64 pKa = 11.84AHH66 pKa = 6.73IEE68 pKa = 3.66VARR71 pKa = 11.84NKK73 pKa = 10.04KK74 pKa = 9.75ALSEE78 pKa = 4.16YY79 pKa = 8.28VHH81 pKa = 7.22KK82 pKa = 10.95SDD84 pKa = 3.81TRR86 pKa = 11.84VAKK89 pKa = 10.7VDD91 pKa = 4.09DD92 pKa = 5.09NISNNIFQWQAEE104 pKa = 4.45VAKK107 pKa = 10.16LWNDD111 pKa = 2.98TDD113 pKa = 4.21FTHH116 pKa = 6.49MKK118 pKa = 10.09YY119 pKa = 10.12IYY121 pKa = 10.15DD122 pKa = 4.07KK123 pKa = 11.08EE124 pKa = 4.16PSEE127 pKa = 4.84DD128 pKa = 3.45VALKK132 pKa = 10.82YY133 pKa = 10.61VDD135 pKa = 4.0SLCGKK140 pKa = 9.91LIRR143 pKa = 11.84EE144 pKa = 4.29GAKK147 pKa = 10.0GLEE150 pKa = 4.21FVAVNPMWRR159 pKa = 11.84SSWKK163 pKa = 9.8RR164 pKa = 11.84FYY166 pKa = 10.99FSIITRR172 pKa = 11.84DD173 pKa = 3.51GRR175 pKa = 11.84DD176 pKa = 3.11EE177 pKa = 4.29AVCEE181 pKa = 4.18TQVPQGEE188 pKa = 4.38EE189 pKa = 4.08TNEE192 pKa = 4.01KK193 pKa = 10.74GSS195 pKa = 3.46
Molecular weight: 22.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA19|A0A126GA19_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-4 OX=1685768 PE=4 SV=1
MM1 pKa = 7.67GGMKK5 pKa = 9.87QYY7 pKa = 11.48AKK9 pKa = 10.29RR10 pKa = 11.84KK11 pKa = 7.23YY12 pKa = 10.11RR13 pKa = 11.84KK14 pKa = 9.28GKK16 pKa = 10.01KK17 pKa = 6.77PTKK20 pKa = 10.0KK21 pKa = 10.11GVKK24 pKa = 9.19INYY27 pKa = 9.19ADD29 pKa = 3.4QHH31 pKa = 5.18NLKK34 pKa = 10.01VQGTSCFIWTSGSYY48 pKa = 10.5PSVQANNALCVTVNPEE64 pKa = 3.98LRR66 pKa = 11.84PPNLSVFAFGGALQFSLGNAINLAALATYY95 pKa = 9.13WDD97 pKa = 4.44RR98 pKa = 11.84IKK100 pKa = 11.22CNEE103 pKa = 3.7IKK105 pKa = 10.82VRR107 pKa = 11.84VIPQINFASVGSGLIPTMRR126 pKa = 11.84TVFDD130 pKa = 4.12FDD132 pKa = 4.73DD133 pKa = 4.22AVTPTSSQMWARR145 pKa = 11.84RR146 pKa = 11.84GKK148 pKa = 6.81THH150 pKa = 7.66RR151 pKa = 11.84LDD153 pKa = 3.51KK154 pKa = 10.96PFTFSFKK161 pKa = 10.52PRR163 pKa = 11.84VLYY166 pKa = 10.32TGVTALSLSQKK177 pKa = 10.07SPYY180 pKa = 9.02MNNASALTIPLYY192 pKa = 10.39GVKK195 pKa = 10.28FGVRR199 pKa = 11.84DD200 pKa = 3.62WPLQAPTDD208 pKa = 4.06PLPNILRR215 pKa = 11.84FEE217 pKa = 4.09ITYY220 pKa = 8.13MCTMKK225 pKa = 10.28EE226 pKa = 4.05QQWINTPVNIADD238 pKa = 3.99YY239 pKa = 10.88GVLLPGQVAPVANEE253 pKa = 3.68VMDD256 pKa = 3.73VSGNIYY262 pKa = 10.32DD263 pKa = 4.87LSGNMIKK270 pKa = 10.67APDD273 pKa = 3.55EE274 pKa = 4.71FGWEE278 pKa = 4.32HH279 pKa = 7.16PWTGPTGLPYY289 pKa = 10.28TGPTGPGPCYY299 pKa = 10.45CC300 pKa = 5.15
MM1 pKa = 7.67GGMKK5 pKa = 9.87QYY7 pKa = 11.48AKK9 pKa = 10.29RR10 pKa = 11.84KK11 pKa = 7.23YY12 pKa = 10.11RR13 pKa = 11.84KK14 pKa = 9.28GKK16 pKa = 10.01KK17 pKa = 6.77PTKK20 pKa = 10.0KK21 pKa = 10.11GVKK24 pKa = 9.19INYY27 pKa = 9.19ADD29 pKa = 3.4QHH31 pKa = 5.18NLKK34 pKa = 10.01VQGTSCFIWTSGSYY48 pKa = 10.5PSVQANNALCVTVNPEE64 pKa = 3.98LRR66 pKa = 11.84PPNLSVFAFGGALQFSLGNAINLAALATYY95 pKa = 9.13WDD97 pKa = 4.44RR98 pKa = 11.84IKK100 pKa = 11.22CNEE103 pKa = 3.7IKK105 pKa = 10.82VRR107 pKa = 11.84VIPQINFASVGSGLIPTMRR126 pKa = 11.84TVFDD130 pKa = 4.12FDD132 pKa = 4.73DD133 pKa = 4.22AVTPTSSQMWARR145 pKa = 11.84RR146 pKa = 11.84GKK148 pKa = 6.81THH150 pKa = 7.66RR151 pKa = 11.84LDD153 pKa = 3.51KK154 pKa = 10.96PFTFSFKK161 pKa = 10.52PRR163 pKa = 11.84VLYY166 pKa = 10.32TGVTALSLSQKK177 pKa = 10.07SPYY180 pKa = 9.02MNNASALTIPLYY192 pKa = 10.39GVKK195 pKa = 10.28FGVRR199 pKa = 11.84DD200 pKa = 3.62WPLQAPTDD208 pKa = 4.06PLPNILRR215 pKa = 11.84FEE217 pKa = 4.09ITYY220 pKa = 8.13MCTMKK225 pKa = 10.28EE226 pKa = 4.05QQWINTPVNIADD238 pKa = 3.99YY239 pKa = 10.88GVLLPGQVAPVANEE253 pKa = 3.68VMDD256 pKa = 3.73VSGNIYY262 pKa = 10.32DD263 pKa = 4.87LSGNMIKK270 pKa = 10.67APDD273 pKa = 3.55EE274 pKa = 4.71FGWEE278 pKa = 4.32HH279 pKa = 7.16PWTGPTGLPYY289 pKa = 10.28TGPTGPGPCYY299 pKa = 10.45CC300 pKa = 5.15
Molecular weight: 33.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
495 |
195 |
300 |
247.5 |
27.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.667 ± 0.319 | 1.818 ± 0.174 |
4.646 ± 0.618 | 4.848 ± 2.406 |
4.04 ± 0.28 | 7.475 ± 0.821 |
1.414 ± 0.396 | 5.253 ± 0.242 |
6.869 ± 0.512 | 6.263 ± 1.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.626 ± 0.358 | 5.455 ± 0.522 |
6.465 ± 2.107 | 4.444 ± 0.425 |
4.444 ± 0.425 | 5.657 ± 0.01 |
7.677 ± 0.329 | 7.273 ± 0.261 |
2.626 ± 0.28 | 4.04 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
