
Lemur associated porprismacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
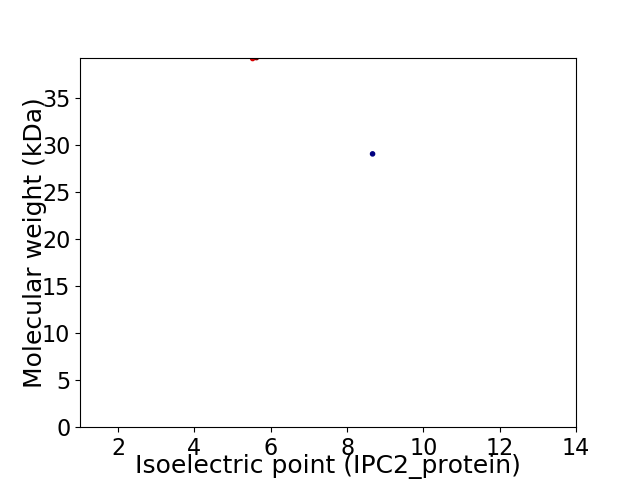
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0B5GP14|A0A0B5GP14_9VIRU Rep protein OS=Lemur associated porprismacovirus 1 OX=2170115 GN=rep PE=4 SV=1
MM1 pKa = 7.83RR2 pKa = 11.84VQISEE7 pKa = 4.39TYY9 pKa = 10.65DD10 pKa = 3.21LSTKK14 pKa = 8.92INKK17 pKa = 8.59MGIVGIHH24 pKa = 5.65TPVGPLLNRR33 pKa = 11.84LYY35 pKa = 10.35PGLVLNYY42 pKa = 10.33KK43 pKa = 9.97KK44 pKa = 10.69FRR46 pKa = 11.84FSKK49 pKa = 10.72CDD51 pKa = 2.92ITLACASMLPADD63 pKa = 4.27PLQVGVEE70 pKa = 4.24AGAIAPQDD78 pKa = 3.66MFNPILFKK86 pKa = 10.81AVSNDD91 pKa = 2.84TMNQFVNYY99 pKa = 7.83IQYY102 pKa = 10.69YY103 pKa = 7.96GASNKK108 pKa = 8.52TLALDD113 pKa = 4.25GSSVVAVNDD122 pKa = 3.71AVFKK126 pKa = 11.03AGSTQATDD134 pKa = 3.34QFSLYY139 pKa = 10.36YY140 pKa = 10.58SLLADD145 pKa = 3.51PKK147 pKa = 9.26GWRR150 pKa = 11.84KK151 pKa = 9.97AMPQSGLQMRR161 pKa = 11.84NLRR164 pKa = 11.84PLVFQVVEE172 pKa = 4.5NGAPLQGRR180 pKa = 11.84VGVADD185 pKa = 4.35PATGDD190 pKa = 3.46INSGAFIGDD199 pKa = 3.77PASGQSYY206 pKa = 8.62TGEE209 pKa = 4.12SWGVPSDD216 pKa = 3.64ASGTAVLSSNKK227 pKa = 9.52GVNIYY232 pKa = 10.1RR233 pKa = 11.84GRR235 pKa = 11.84AIRR238 pKa = 11.84MPWISTKK245 pKa = 10.25FFNNGTGVADD255 pKa = 4.45SNTIMPSFAQIVEE268 pKa = 4.27GTNVSTNTGRR278 pKa = 11.84VPPTYY283 pKa = 9.19VACIIVPPAKK293 pKa = 10.61LNVLYY298 pKa = 11.08YY299 pKa = 10.4RR300 pKa = 11.84MKK302 pKa = 9.73VTWTVEE308 pKa = 4.08FTGLSSMTPYY318 pKa = 10.84LGWNDD323 pKa = 3.61LTDD326 pKa = 3.85TADD329 pKa = 2.99IAYY332 pKa = 7.57GTDD335 pKa = 3.31YY336 pKa = 11.66ANQASVAMTSLEE348 pKa = 4.24TMVDD352 pKa = 3.26TSEE355 pKa = 4.21VDD357 pKa = 2.94MSKK360 pKa = 10.44IMEE363 pKa = 4.23GG364 pKa = 3.32
MM1 pKa = 7.83RR2 pKa = 11.84VQISEE7 pKa = 4.39TYY9 pKa = 10.65DD10 pKa = 3.21LSTKK14 pKa = 8.92INKK17 pKa = 8.59MGIVGIHH24 pKa = 5.65TPVGPLLNRR33 pKa = 11.84LYY35 pKa = 10.35PGLVLNYY42 pKa = 10.33KK43 pKa = 9.97KK44 pKa = 10.69FRR46 pKa = 11.84FSKK49 pKa = 10.72CDD51 pKa = 2.92ITLACASMLPADD63 pKa = 4.27PLQVGVEE70 pKa = 4.24AGAIAPQDD78 pKa = 3.66MFNPILFKK86 pKa = 10.81AVSNDD91 pKa = 2.84TMNQFVNYY99 pKa = 7.83IQYY102 pKa = 10.69YY103 pKa = 7.96GASNKK108 pKa = 8.52TLALDD113 pKa = 4.25GSSVVAVNDD122 pKa = 3.71AVFKK126 pKa = 11.03AGSTQATDD134 pKa = 3.34QFSLYY139 pKa = 10.36YY140 pKa = 10.58SLLADD145 pKa = 3.51PKK147 pKa = 9.26GWRR150 pKa = 11.84KK151 pKa = 9.97AMPQSGLQMRR161 pKa = 11.84NLRR164 pKa = 11.84PLVFQVVEE172 pKa = 4.5NGAPLQGRR180 pKa = 11.84VGVADD185 pKa = 4.35PATGDD190 pKa = 3.46INSGAFIGDD199 pKa = 3.77PASGQSYY206 pKa = 8.62TGEE209 pKa = 4.12SWGVPSDD216 pKa = 3.64ASGTAVLSSNKK227 pKa = 9.52GVNIYY232 pKa = 10.1RR233 pKa = 11.84GRR235 pKa = 11.84AIRR238 pKa = 11.84MPWISTKK245 pKa = 10.25FFNNGTGVADD255 pKa = 4.45SNTIMPSFAQIVEE268 pKa = 4.27GTNVSTNTGRR278 pKa = 11.84VPPTYY283 pKa = 9.19VACIIVPPAKK293 pKa = 10.61LNVLYY298 pKa = 11.08YY299 pKa = 10.4RR300 pKa = 11.84MKK302 pKa = 9.73VTWTVEE308 pKa = 4.08FTGLSSMTPYY318 pKa = 10.84LGWNDD323 pKa = 3.61LTDD326 pKa = 3.85TADD329 pKa = 2.99IAYY332 pKa = 7.57GTDD335 pKa = 3.31YY336 pKa = 11.66ANQASVAMTSLEE348 pKa = 4.24TMVDD352 pKa = 3.26TSEE355 pKa = 4.21VDD357 pKa = 2.94MSKK360 pKa = 10.44IMEE363 pKa = 4.23GG364 pKa = 3.32
Molecular weight: 39.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5GP14|A0A0B5GP14_9VIRU Rep protein OS=Lemur associated porprismacovirus 1 OX=2170115 GN=rep PE=4 SV=1
MM1 pKa = 7.21TNVYY5 pKa = 9.44MITMPRR11 pKa = 11.84KK12 pKa = 9.07GYY14 pKa = 10.4SGFIAIMRR22 pKa = 11.84FFRR25 pKa = 11.84KK26 pKa = 9.61FDD28 pKa = 3.26VHH30 pKa = 6.97KK31 pKa = 10.34WIVSPEE37 pKa = 3.9KK38 pKa = 10.01GAQGYY43 pKa = 7.27EE44 pKa = 3.37HH45 pKa = 6.01WQIRR49 pKa = 11.84FRR51 pKa = 11.84CGLTPEE57 pKa = 4.2NAMIAWRR64 pKa = 11.84QWVCNGFNMLEE75 pKa = 4.09ASDD78 pKa = 4.79NGWEE82 pKa = 4.08YY83 pKa = 10.96EE84 pKa = 4.13GKK86 pKa = 9.97EE87 pKa = 4.29GKK89 pKa = 9.86FLASWDD95 pKa = 3.92SKK97 pKa = 9.87GARR100 pKa = 11.84SVRR103 pKa = 11.84FGKK106 pKa = 9.07MEE108 pKa = 3.63WRR110 pKa = 11.84QEE112 pKa = 4.02ATVLRR117 pKa = 11.84ARR119 pKa = 11.84ATNDD123 pKa = 2.97RR124 pKa = 11.84EE125 pKa = 4.06IVVWYY130 pKa = 9.72DD131 pKa = 3.16PKK133 pKa = 11.42GNSGKK138 pKa = 9.99SWLVGHH144 pKa = 6.86LVEE147 pKa = 4.86TRR149 pKa = 11.84QAYY152 pKa = 7.69YY153 pKa = 10.42VPPYY157 pKa = 9.89LATVEE162 pKa = 4.82SMIKK166 pKa = 9.11TLASMVKK173 pKa = 10.14ADD175 pKa = 4.83RR176 pKa = 11.84EE177 pKa = 4.32NGNPPRR183 pKa = 11.84PLVVIDD189 pKa = 3.94IPRR192 pKa = 11.84SYY194 pKa = 10.36KK195 pKa = 9.16WKK197 pKa = 9.38NDD199 pKa = 3.0MYY201 pKa = 11.33VAIEE205 pKa = 4.42AIKK208 pKa = 10.62DD209 pKa = 3.88GIIVDD214 pKa = 3.85PRR216 pKa = 11.84YY217 pKa = 10.12SATVEE222 pKa = 3.95NVKK225 pKa = 10.88GIGVIVITNEE235 pKa = 3.49KK236 pKa = 8.57PQVGKK241 pKa = 10.76LSADD245 pKa = 2.98RR246 pKa = 11.84WDD248 pKa = 3.48IVDD251 pKa = 3.32WW252 pKa = 4.39
MM1 pKa = 7.21TNVYY5 pKa = 9.44MITMPRR11 pKa = 11.84KK12 pKa = 9.07GYY14 pKa = 10.4SGFIAIMRR22 pKa = 11.84FFRR25 pKa = 11.84KK26 pKa = 9.61FDD28 pKa = 3.26VHH30 pKa = 6.97KK31 pKa = 10.34WIVSPEE37 pKa = 3.9KK38 pKa = 10.01GAQGYY43 pKa = 7.27EE44 pKa = 3.37HH45 pKa = 6.01WQIRR49 pKa = 11.84FRR51 pKa = 11.84CGLTPEE57 pKa = 4.2NAMIAWRR64 pKa = 11.84QWVCNGFNMLEE75 pKa = 4.09ASDD78 pKa = 4.79NGWEE82 pKa = 4.08YY83 pKa = 10.96EE84 pKa = 4.13GKK86 pKa = 9.97EE87 pKa = 4.29GKK89 pKa = 9.86FLASWDD95 pKa = 3.92SKK97 pKa = 9.87GARR100 pKa = 11.84SVRR103 pKa = 11.84FGKK106 pKa = 9.07MEE108 pKa = 3.63WRR110 pKa = 11.84QEE112 pKa = 4.02ATVLRR117 pKa = 11.84ARR119 pKa = 11.84ATNDD123 pKa = 2.97RR124 pKa = 11.84EE125 pKa = 4.06IVVWYY130 pKa = 9.72DD131 pKa = 3.16PKK133 pKa = 11.42GNSGKK138 pKa = 9.99SWLVGHH144 pKa = 6.86LVEE147 pKa = 4.86TRR149 pKa = 11.84QAYY152 pKa = 7.69YY153 pKa = 10.42VPPYY157 pKa = 9.89LATVEE162 pKa = 4.82SMIKK166 pKa = 9.11TLASMVKK173 pKa = 10.14ADD175 pKa = 4.83RR176 pKa = 11.84EE177 pKa = 4.32NGNPPRR183 pKa = 11.84PLVVIDD189 pKa = 3.94IPRR192 pKa = 11.84SYY194 pKa = 10.36KK195 pKa = 9.16WKK197 pKa = 9.38NDD199 pKa = 3.0MYY201 pKa = 11.33VAIEE205 pKa = 4.42AIKK208 pKa = 10.62DD209 pKa = 3.88GIIVDD214 pKa = 3.85PRR216 pKa = 11.84YY217 pKa = 10.12SATVEE222 pKa = 3.95NVKK225 pKa = 10.88GIGVIVITNEE235 pKa = 3.49KK236 pKa = 8.57PQVGKK241 pKa = 10.76LSADD245 pKa = 2.98RR246 pKa = 11.84WDD248 pKa = 3.48IVDD251 pKa = 3.32WW252 pKa = 4.39
Molecular weight: 29.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
616 |
252 |
364 |
308.0 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.117 ± 0.584 | 0.812 ± 0.011 |
5.357 ± 0.119 | 4.058 ± 1.375 |
3.409 ± 0.141 | 8.117 ± 0.346 |
0.649 ± 0.325 | 6.006 ± 0.682 |
5.357 ± 1.072 | 5.844 ± 1.126 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.058 ± 0.054 | 5.519 ± 0.455 |
5.357 ± 0.357 | 3.247 ± 0.52 |
4.87 ± 1.364 | 6.981 ± 1.093 |
6.169 ± 1.32 | 8.929 ± 0.119 |
2.76 ± 1.201 | 4.383 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
