
Entoleuca phenui-like virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Entovirus; Entoleuca entovirus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
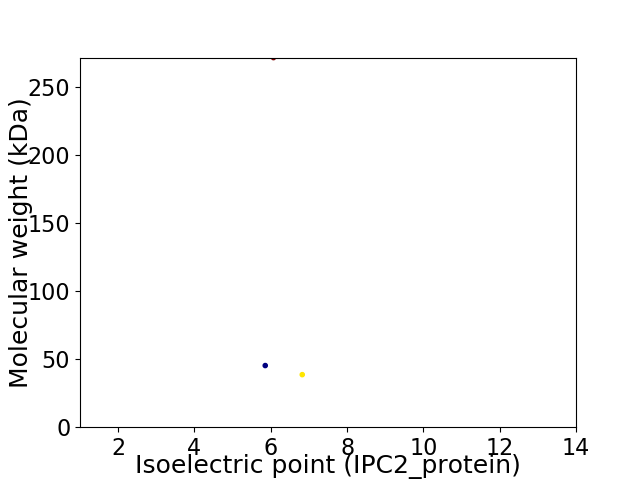
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A4D6Q3I0|A0A4D6Q3I0_9VIRU Movement protein OS=Entoleuca phenui-like virus 1 OX=2086640 PE=4 SV=1
MM1 pKa = 7.38LEE3 pKa = 4.19SVKK6 pKa = 10.04MLRR9 pKa = 11.84KK10 pKa = 9.36FFGRR14 pKa = 11.84RR15 pKa = 11.84GSDD18 pKa = 3.14NKK20 pKa = 10.61PSGARR25 pKa = 11.84PGGSIRR31 pKa = 11.84GSITGSRR38 pKa = 11.84KK39 pKa = 9.25MKK41 pKa = 10.61DD42 pKa = 2.87KK43 pKa = 9.98EE44 pKa = 4.53TEE46 pKa = 3.72DD47 pKa = 3.51RR48 pKa = 11.84EE49 pKa = 4.32ARR51 pKa = 11.84RR52 pKa = 11.84GAVLRR57 pKa = 11.84NMDD60 pKa = 3.74EE61 pKa = 4.28ALFASGVPKK70 pKa = 10.58NHH72 pKa = 5.84VSMADD77 pKa = 3.13KK78 pKa = 10.82LDD80 pKa = 3.51SHH82 pKa = 6.94KK83 pKa = 11.14AGSTFKK89 pKa = 10.61YY90 pKa = 10.41AVIGTQPSEE99 pKa = 3.9EE100 pKa = 3.94RR101 pKa = 11.84TGIMNKK107 pKa = 9.39LRR109 pKa = 11.84NYY111 pKa = 8.8KK112 pKa = 10.41APSKK116 pKa = 10.21LQNFSLSDD124 pKa = 3.27IPFIRR129 pKa = 11.84LMDD132 pKa = 4.33DD133 pKa = 3.19EE134 pKa = 5.81VEE136 pKa = 4.5FSMKK140 pKa = 10.03EE141 pKa = 3.72ALRR144 pKa = 11.84SSNAQKK150 pKa = 10.86HH151 pKa = 5.15EE152 pKa = 4.44DD153 pKa = 3.56YY154 pKa = 11.33CLIDD158 pKa = 3.23SVLIHH163 pKa = 6.25FVPLDD168 pKa = 3.31SFLNDD173 pKa = 3.76KK174 pKa = 11.19SPITIQLNDD183 pKa = 3.42FRR185 pKa = 11.84RR186 pKa = 11.84VNNTVARR193 pKa = 11.84LATVDD198 pKa = 3.12NTMSYY203 pKa = 11.16NILFCLDD210 pKa = 3.48YY211 pKa = 10.95CVEE214 pKa = 4.12SRR216 pKa = 11.84DD217 pKa = 3.97LEE219 pKa = 4.3RR220 pKa = 11.84MTLSFACPQKK230 pKa = 10.81DD231 pKa = 3.85FQPGVAWGAVKK242 pKa = 10.45VIAQVKK248 pKa = 9.25FMSFPVRR255 pKa = 11.84MPVIEE260 pKa = 4.23TMAVAIFADD269 pKa = 3.94TDD271 pKa = 3.92LLEE274 pKa = 4.48YY275 pKa = 10.3EE276 pKa = 3.87WDD278 pKa = 3.53PRR280 pKa = 11.84EE281 pKa = 3.92FDD283 pKa = 4.41AVLGPQALQGVRR295 pKa = 11.84QANKK299 pKa = 10.06RR300 pKa = 11.84GEE302 pKa = 4.18LEE304 pKa = 3.88NLTIAKK310 pKa = 9.1NDD312 pKa = 3.34RR313 pKa = 11.84MEE315 pKa = 4.14VGMARR320 pKa = 11.84TLAMGSLEE328 pKa = 3.99EE329 pKa = 4.46EE330 pKa = 4.51EE331 pKa = 4.46PGEE334 pKa = 4.98AIQALKK340 pKa = 10.62NAALLRR346 pKa = 11.84QRR348 pKa = 11.84QANSTGKK355 pKa = 10.59GKK357 pKa = 10.99GILKK361 pKa = 9.87HH362 pKa = 7.03DD363 pKa = 4.36SFDD366 pKa = 3.83SPTEE370 pKa = 4.0EE371 pKa = 5.53KK372 pKa = 10.76EE373 pKa = 4.24LDD375 pKa = 3.88PDD377 pKa = 4.21DD378 pKa = 4.77SASQVLSGLSEE389 pKa = 4.82EE390 pKa = 4.6IPGPRR395 pKa = 11.84NVTGSRR401 pKa = 11.84TVNFTT406 pKa = 3.2
MM1 pKa = 7.38LEE3 pKa = 4.19SVKK6 pKa = 10.04MLRR9 pKa = 11.84KK10 pKa = 9.36FFGRR14 pKa = 11.84RR15 pKa = 11.84GSDD18 pKa = 3.14NKK20 pKa = 10.61PSGARR25 pKa = 11.84PGGSIRR31 pKa = 11.84GSITGSRR38 pKa = 11.84KK39 pKa = 9.25MKK41 pKa = 10.61DD42 pKa = 2.87KK43 pKa = 9.98EE44 pKa = 4.53TEE46 pKa = 3.72DD47 pKa = 3.51RR48 pKa = 11.84EE49 pKa = 4.32ARR51 pKa = 11.84RR52 pKa = 11.84GAVLRR57 pKa = 11.84NMDD60 pKa = 3.74EE61 pKa = 4.28ALFASGVPKK70 pKa = 10.58NHH72 pKa = 5.84VSMADD77 pKa = 3.13KK78 pKa = 10.82LDD80 pKa = 3.51SHH82 pKa = 6.94KK83 pKa = 11.14AGSTFKK89 pKa = 10.61YY90 pKa = 10.41AVIGTQPSEE99 pKa = 3.9EE100 pKa = 3.94RR101 pKa = 11.84TGIMNKK107 pKa = 9.39LRR109 pKa = 11.84NYY111 pKa = 8.8KK112 pKa = 10.41APSKK116 pKa = 10.21LQNFSLSDD124 pKa = 3.27IPFIRR129 pKa = 11.84LMDD132 pKa = 4.33DD133 pKa = 3.19EE134 pKa = 5.81VEE136 pKa = 4.5FSMKK140 pKa = 10.03EE141 pKa = 3.72ALRR144 pKa = 11.84SSNAQKK150 pKa = 10.86HH151 pKa = 5.15EE152 pKa = 4.44DD153 pKa = 3.56YY154 pKa = 11.33CLIDD158 pKa = 3.23SVLIHH163 pKa = 6.25FVPLDD168 pKa = 3.31SFLNDD173 pKa = 3.76KK174 pKa = 11.19SPITIQLNDD183 pKa = 3.42FRR185 pKa = 11.84RR186 pKa = 11.84VNNTVARR193 pKa = 11.84LATVDD198 pKa = 3.12NTMSYY203 pKa = 11.16NILFCLDD210 pKa = 3.48YY211 pKa = 10.95CVEE214 pKa = 4.12SRR216 pKa = 11.84DD217 pKa = 3.97LEE219 pKa = 4.3RR220 pKa = 11.84MTLSFACPQKK230 pKa = 10.81DD231 pKa = 3.85FQPGVAWGAVKK242 pKa = 10.45VIAQVKK248 pKa = 9.25FMSFPVRR255 pKa = 11.84MPVIEE260 pKa = 4.23TMAVAIFADD269 pKa = 3.94TDD271 pKa = 3.92LLEE274 pKa = 4.48YY275 pKa = 10.3EE276 pKa = 3.87WDD278 pKa = 3.53PRR280 pKa = 11.84EE281 pKa = 3.92FDD283 pKa = 4.41AVLGPQALQGVRR295 pKa = 11.84QANKK299 pKa = 10.06RR300 pKa = 11.84GEE302 pKa = 4.18LEE304 pKa = 3.88NLTIAKK310 pKa = 9.1NDD312 pKa = 3.34RR313 pKa = 11.84MEE315 pKa = 4.14VGMARR320 pKa = 11.84TLAMGSLEE328 pKa = 3.99EE329 pKa = 4.46EE330 pKa = 4.51EE331 pKa = 4.46PGEE334 pKa = 4.98AIQALKK340 pKa = 10.62NAALLRR346 pKa = 11.84QRR348 pKa = 11.84QANSTGKK355 pKa = 10.59GKK357 pKa = 10.99GILKK361 pKa = 9.87HH362 pKa = 7.03DD363 pKa = 4.36SFDD366 pKa = 3.83SPTEE370 pKa = 4.0EE371 pKa = 5.53KK372 pKa = 10.76EE373 pKa = 4.24LDD375 pKa = 3.88PDD377 pKa = 4.21DD378 pKa = 4.77SASQVLSGLSEE389 pKa = 4.82EE390 pKa = 4.6IPGPRR395 pKa = 11.84NVTGSRR401 pKa = 11.84TVNFTT406 pKa = 3.2
Molecular weight: 45.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6Q3I0|A0A4D6Q3I0_9VIRU Movement protein OS=Entoleuca phenui-like virus 1 OX=2086640 PE=4 SV=1
MM1 pKa = 7.39SSSSRR6 pKa = 11.84KK7 pKa = 5.44VTGVGGQGQGSSGGPAQGGSQAPSISGMVDD37 pKa = 2.69KK38 pKa = 10.96LGNKK42 pKa = 8.7IWLTNTSTSEE52 pKa = 4.03DD53 pKa = 3.56LTPAKK58 pKa = 10.12ISEE61 pKa = 4.24LVQNVSNDD69 pKa = 4.34DD70 pKa = 3.16ISKK73 pKa = 9.47TITAVEE79 pKa = 4.33SSTTTARR86 pKa = 11.84DD87 pKa = 3.24ALEE90 pKa = 3.88FSIFDD95 pKa = 3.72FQGFDD100 pKa = 3.44PLMVWRR106 pKa = 11.84VLRR109 pKa = 11.84ACQSYY114 pKa = 10.95YY115 pKa = 10.67KK116 pKa = 10.87DD117 pKa = 3.2SDD119 pKa = 3.76EE120 pKa = 5.03NLLSDD125 pKa = 4.02IRR127 pKa = 11.84FSVAAVLYY135 pKa = 9.23MGNLQTKK142 pKa = 9.25ALTRR146 pKa = 11.84RR147 pKa = 11.84AQEE150 pKa = 3.52GRR152 pKa = 11.84TKK154 pKa = 10.18IEE156 pKa = 4.03YY157 pKa = 9.74LVKK160 pKa = 10.3KK161 pKa = 9.95YY162 pKa = 10.8NIRR165 pKa = 11.84TGSSGAGLEE174 pKa = 4.33AEE176 pKa = 4.36TLTFPRR182 pKa = 11.84IAAAAPVFAVKK193 pKa = 10.22LANQLPPSAVSIAFMGRR210 pKa = 11.84EE211 pKa = 4.22VPSPMRR217 pKa = 11.84LMPFASLCSPQMDD230 pKa = 3.73TEE232 pKa = 4.13LRR234 pKa = 11.84KK235 pKa = 9.5FLLQACNAHH244 pKa = 6.33ASDD247 pKa = 3.72MGLAYY252 pKa = 10.05EE253 pKa = 5.14RR254 pKa = 11.84GRR256 pKa = 11.84CKK258 pKa = 10.11KK259 pKa = 10.17AKK261 pKa = 9.93KK262 pKa = 10.17EE263 pKa = 3.9FVANPKK269 pKa = 9.98EE270 pKa = 3.95MAEE273 pKa = 4.22DD274 pKa = 3.03QWAFIEE280 pKa = 4.46VASDD284 pKa = 3.64SPVPEE289 pKa = 4.15EE290 pKa = 3.96SMKK293 pKa = 10.75RR294 pKa = 11.84STLLEE299 pKa = 4.55LNLSQYY305 pKa = 10.4YY306 pKa = 9.35KK307 pKa = 10.57QLAEE311 pKa = 3.93VVRR314 pKa = 11.84NYY316 pKa = 10.65RR317 pKa = 11.84SIMEE321 pKa = 4.28GSEE324 pKa = 3.85KK325 pKa = 10.23STVKK329 pKa = 10.62VMSQSEE335 pKa = 4.42FDD337 pKa = 3.5TKK339 pKa = 10.87LSEE342 pKa = 4.97FISSRR347 pKa = 11.84AMDD350 pKa = 3.66TQQ352 pKa = 3.31
MM1 pKa = 7.39SSSSRR6 pKa = 11.84KK7 pKa = 5.44VTGVGGQGQGSSGGPAQGGSQAPSISGMVDD37 pKa = 2.69KK38 pKa = 10.96LGNKK42 pKa = 8.7IWLTNTSTSEE52 pKa = 4.03DD53 pKa = 3.56LTPAKK58 pKa = 10.12ISEE61 pKa = 4.24LVQNVSNDD69 pKa = 4.34DD70 pKa = 3.16ISKK73 pKa = 9.47TITAVEE79 pKa = 4.33SSTTTARR86 pKa = 11.84DD87 pKa = 3.24ALEE90 pKa = 3.88FSIFDD95 pKa = 3.72FQGFDD100 pKa = 3.44PLMVWRR106 pKa = 11.84VLRR109 pKa = 11.84ACQSYY114 pKa = 10.95YY115 pKa = 10.67KK116 pKa = 10.87DD117 pKa = 3.2SDD119 pKa = 3.76EE120 pKa = 5.03NLLSDD125 pKa = 4.02IRR127 pKa = 11.84FSVAAVLYY135 pKa = 9.23MGNLQTKK142 pKa = 9.25ALTRR146 pKa = 11.84RR147 pKa = 11.84AQEE150 pKa = 3.52GRR152 pKa = 11.84TKK154 pKa = 10.18IEE156 pKa = 4.03YY157 pKa = 9.74LVKK160 pKa = 10.3KK161 pKa = 9.95YY162 pKa = 10.8NIRR165 pKa = 11.84TGSSGAGLEE174 pKa = 4.33AEE176 pKa = 4.36TLTFPRR182 pKa = 11.84IAAAAPVFAVKK193 pKa = 10.22LANQLPPSAVSIAFMGRR210 pKa = 11.84EE211 pKa = 4.22VPSPMRR217 pKa = 11.84LMPFASLCSPQMDD230 pKa = 3.73TEE232 pKa = 4.13LRR234 pKa = 11.84KK235 pKa = 9.5FLLQACNAHH244 pKa = 6.33ASDD247 pKa = 3.72MGLAYY252 pKa = 10.05EE253 pKa = 5.14RR254 pKa = 11.84GRR256 pKa = 11.84CKK258 pKa = 10.11KK259 pKa = 10.17AKK261 pKa = 9.93KK262 pKa = 10.17EE263 pKa = 3.9FVANPKK269 pKa = 9.98EE270 pKa = 3.95MAEE273 pKa = 4.22DD274 pKa = 3.03QWAFIEE280 pKa = 4.46VASDD284 pKa = 3.64SPVPEE289 pKa = 4.15EE290 pKa = 3.96SMKK293 pKa = 10.75RR294 pKa = 11.84STLLEE299 pKa = 4.55LNLSQYY305 pKa = 10.4YY306 pKa = 9.35KK307 pKa = 10.57QLAEE311 pKa = 3.93VVRR314 pKa = 11.84NYY316 pKa = 10.65RR317 pKa = 11.84SIMEE321 pKa = 4.28GSEE324 pKa = 3.85KK325 pKa = 10.23STVKK329 pKa = 10.62VMSQSEE335 pKa = 4.42FDD337 pKa = 3.5TKK339 pKa = 10.87LSEE342 pKa = 4.97FISSRR347 pKa = 11.84AMDD350 pKa = 3.66TQQ352 pKa = 3.31
Molecular weight: 38.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3111 |
352 |
2353 |
1037.0 |
118.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.625 ± 1.479 | 1.382 ± 0.16 |
6.107 ± 0.426 | 7.49 ± 0.208 |
4.114 ± 0.158 | 5.239 ± 0.469 |
2.089 ± 0.664 | 5.947 ± 0.851 |
7.425 ± 0.536 | 9.129 ± 0.345 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.6 ± 0.175 | 4.629 ± 0.331 |
3.729 ± 0.37 | 3.632 ± 0.331 |
5.561 ± 0.456 | 8.454 ± 0.978 |
4.982 ± 0.265 | 6.011 ± 0.095 |
1.446 ± 0.384 | 3.407 ± 0.706 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
