
Moumouvirus australiensis
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Imitervirales; Mimiviridae; unclassified Mimiviridae; Moumouvirus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 882 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2P1EM83|A0A2P1EM83_9VIRU Uncharacterized protein OS=Moumouvirus australiensis OX=2109587 GN=mc_619 PE=4 SV=1
MM1 pKa = 7.44GNSTSTNTQTLEE13 pKa = 3.93NQPLINSVSEE23 pKa = 4.61DD24 pKa = 3.67NVLMNYY30 pKa = 9.64QSSTEE35 pKa = 4.11TNEE38 pKa = 3.44EE39 pKa = 4.08VIPVKK44 pKa = 10.28KK45 pKa = 10.23SSYY48 pKa = 9.33IHH50 pKa = 6.4PLINLITVNTTDD62 pKa = 5.22DD63 pKa = 5.21DD64 pKa = 5.39IIDD67 pKa = 4.03TFNAYY72 pKa = 10.32CGVQKK77 pKa = 10.99LSDD80 pKa = 4.03NLYY83 pKa = 10.25DD84 pKa = 4.84IPDD87 pKa = 3.42QYY89 pKa = 11.83EE90 pKa = 3.91VFGPVMQLFCHH101 pKa = 6.25CACYY105 pKa = 10.31GRR107 pKa = 11.84KK108 pKa = 9.22KK109 pKa = 10.32VVDD112 pKa = 4.69WIINNYY118 pKa = 9.2VPLDD122 pKa = 3.5VSYY125 pKa = 11.54SDD127 pKa = 3.2NFCYY131 pKa = 10.35FEE133 pKa = 4.11CLRR136 pKa = 11.84YY137 pKa = 9.31GHH139 pKa = 6.77KK140 pKa = 10.52SIAAMLIQHH149 pKa = 6.47EE150 pKa = 4.76SFNGSLDD157 pKa = 3.58VLLDD161 pKa = 3.48LLNRR165 pKa = 11.84QEE167 pKa = 4.22YY168 pKa = 9.73NQFTTCVNKK177 pKa = 10.54NNNLDD182 pKa = 3.87EE183 pKa = 4.13NLKK186 pKa = 10.47NIRR189 pKa = 11.84STLVYY194 pKa = 10.25LVNTGDD200 pKa = 3.63VQKK203 pKa = 10.88VKK205 pKa = 10.88SILTEE210 pKa = 3.96YY211 pKa = 10.91KK212 pKa = 8.22STKK215 pKa = 10.1VINTEE220 pKa = 3.92SLSQIDD226 pKa = 3.7ITQSTSNIVSVSDD239 pKa = 3.56EE240 pKa = 5.13PITVDD245 pKa = 4.64LGVEE249 pKa = 4.21QPVEE253 pKa = 4.11QPTEE257 pKa = 3.95QPTEE261 pKa = 3.86QSTEE265 pKa = 3.91QPTEE269 pKa = 3.8QPTEE273 pKa = 3.86QSTEE277 pKa = 3.88QPTEE281 pKa = 3.65QSTEE285 pKa = 3.88QPTEE289 pKa = 3.62QSTVQLTEE297 pKa = 3.73QPTEE301 pKa = 3.78QLTEE305 pKa = 3.88QLTEE309 pKa = 3.85QLTEE313 pKa = 3.92QLTEE317 pKa = 4.05QPVEE321 pKa = 4.17QPVEE325 pKa = 4.14QPVEE329 pKa = 4.1QPTEE333 pKa = 4.01QPVEE337 pKa = 4.08QPTEE341 pKa = 3.85QLTEE345 pKa = 4.04QPVEE349 pKa = 4.16QPTEE353 pKa = 3.9QPIGVINPDD362 pKa = 2.97ISIDD366 pKa = 4.12PIQQ369 pKa = 4.23
MM1 pKa = 7.44GNSTSTNTQTLEE13 pKa = 3.93NQPLINSVSEE23 pKa = 4.61DD24 pKa = 3.67NVLMNYY30 pKa = 9.64QSSTEE35 pKa = 4.11TNEE38 pKa = 3.44EE39 pKa = 4.08VIPVKK44 pKa = 10.28KK45 pKa = 10.23SSYY48 pKa = 9.33IHH50 pKa = 6.4PLINLITVNTTDD62 pKa = 5.22DD63 pKa = 5.21DD64 pKa = 5.39IIDD67 pKa = 4.03TFNAYY72 pKa = 10.32CGVQKK77 pKa = 10.99LSDD80 pKa = 4.03NLYY83 pKa = 10.25DD84 pKa = 4.84IPDD87 pKa = 3.42QYY89 pKa = 11.83EE90 pKa = 3.91VFGPVMQLFCHH101 pKa = 6.25CACYY105 pKa = 10.31GRR107 pKa = 11.84KK108 pKa = 9.22KK109 pKa = 10.32VVDD112 pKa = 4.69WIINNYY118 pKa = 9.2VPLDD122 pKa = 3.5VSYY125 pKa = 11.54SDD127 pKa = 3.2NFCYY131 pKa = 10.35FEE133 pKa = 4.11CLRR136 pKa = 11.84YY137 pKa = 9.31GHH139 pKa = 6.77KK140 pKa = 10.52SIAAMLIQHH149 pKa = 6.47EE150 pKa = 4.76SFNGSLDD157 pKa = 3.58VLLDD161 pKa = 3.48LLNRR165 pKa = 11.84QEE167 pKa = 4.22YY168 pKa = 9.73NQFTTCVNKK177 pKa = 10.54NNNLDD182 pKa = 3.87EE183 pKa = 4.13NLKK186 pKa = 10.47NIRR189 pKa = 11.84STLVYY194 pKa = 10.25LVNTGDD200 pKa = 3.63VQKK203 pKa = 10.88VKK205 pKa = 10.88SILTEE210 pKa = 3.96YY211 pKa = 10.91KK212 pKa = 8.22STKK215 pKa = 10.1VINTEE220 pKa = 3.92SLSQIDD226 pKa = 3.7ITQSTSNIVSVSDD239 pKa = 3.56EE240 pKa = 5.13PITVDD245 pKa = 4.64LGVEE249 pKa = 4.21QPVEE253 pKa = 4.11QPTEE257 pKa = 3.95QPTEE261 pKa = 3.86QSTEE265 pKa = 3.91QPTEE269 pKa = 3.8QPTEE273 pKa = 3.86QSTEE277 pKa = 3.88QPTEE281 pKa = 3.65QSTEE285 pKa = 3.88QPTEE289 pKa = 3.62QSTVQLTEE297 pKa = 3.73QPTEE301 pKa = 3.78QLTEE305 pKa = 3.88QLTEE309 pKa = 3.85QLTEE313 pKa = 3.92QLTEE317 pKa = 4.05QPVEE321 pKa = 4.17QPVEE325 pKa = 4.14QPVEE329 pKa = 4.1QPTEE333 pKa = 4.01QPVEE337 pKa = 4.08QPTEE341 pKa = 3.85QLTEE345 pKa = 4.04QPVEE349 pKa = 4.16QPTEE353 pKa = 3.9QPIGVINPDD362 pKa = 2.97ISIDD366 pKa = 4.12PIQQ369 pKa = 4.23
Molecular weight: 41.73 kDa
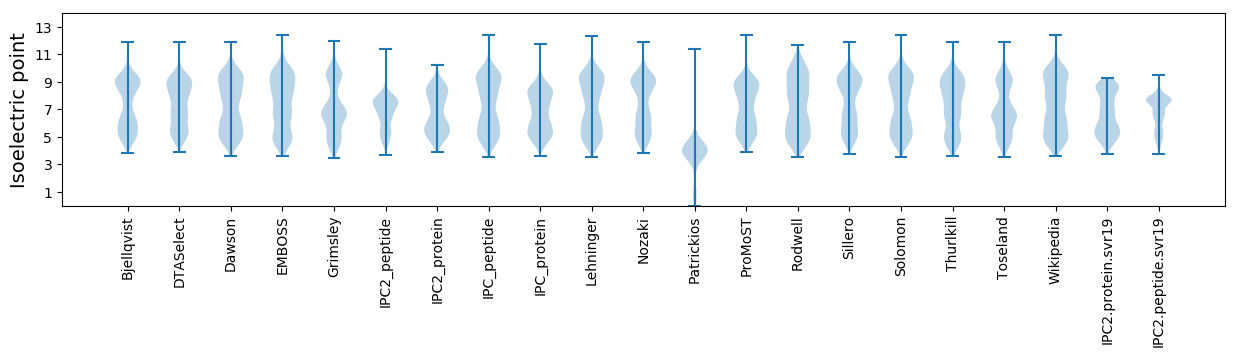
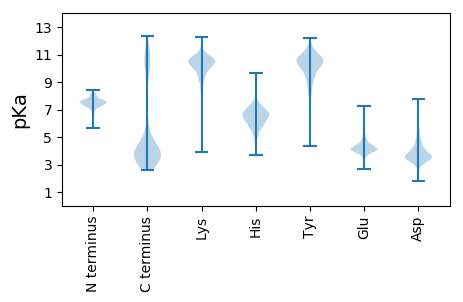
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1EKT4|A0A2P1EKT4_9VIRU Uncharacterized protein OS=Moumouvirus australiensis OX=2109587 GN=mc_117 PE=4 SV=1
MM1 pKa = 7.23NRR3 pKa = 11.84NIMFARR9 pKa = 11.84RR10 pKa = 11.84SSKK13 pKa = 8.83FTSIISTQGFNNMKK27 pKa = 8.3TQYY30 pKa = 10.24SRR32 pKa = 11.84SIYY35 pKa = 7.84TRR37 pKa = 11.84RR38 pKa = 11.84NTNINKK44 pKa = 9.56KK45 pKa = 10.52VIIEE49 pKa = 4.19SLDD52 pKa = 3.37SFMDD56 pKa = 4.43GFTNGCLFGSVVILCYY72 pKa = 10.62INMM75 pKa = 4.44
MM1 pKa = 7.23NRR3 pKa = 11.84NIMFARR9 pKa = 11.84RR10 pKa = 11.84SSKK13 pKa = 8.83FTSIISTQGFNNMKK27 pKa = 8.3TQYY30 pKa = 10.24SRR32 pKa = 11.84SIYY35 pKa = 7.84TRR37 pKa = 11.84RR38 pKa = 11.84NTNINKK44 pKa = 9.56KK45 pKa = 10.52VIIEE49 pKa = 4.19SLDD52 pKa = 3.37SFMDD56 pKa = 4.43GFTNGCLFGSVVILCYY72 pKa = 10.62INMM75 pKa = 4.44
Molecular weight: 8.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
314460 |
64 |
3382 |
356.5 |
41.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.971 ± 0.082 | 1.805 ± 0.053 |
6.463 ± 0.12 | 5.683 ± 0.089 |
4.841 ± 0.069 | 5.062 ± 0.319 |
1.974 ± 0.047 | 10.246 ± 0.129 |
9.29 ± 0.122 | 8.351 ± 0.096 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.865 ± 0.048 | 9.624 ± 0.138 |
3.297 ± 0.075 | 3.024 ± 0.059 |
2.95 ± 0.059 | 6.651 ± 0.084 |
4.884 ± 0.058 | 4.579 ± 0.066 |
0.825 ± 0.031 | 5.615 ± 0.098 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
