
Desulfovibrio salexigens (strain ATCC 14822 / DSM 2638 / NCIMB 8403 / VKM B-1763) (Maridesulfovibrio salexigens)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Maridesulfovibrio; Maridesulfovibrio salexigens
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
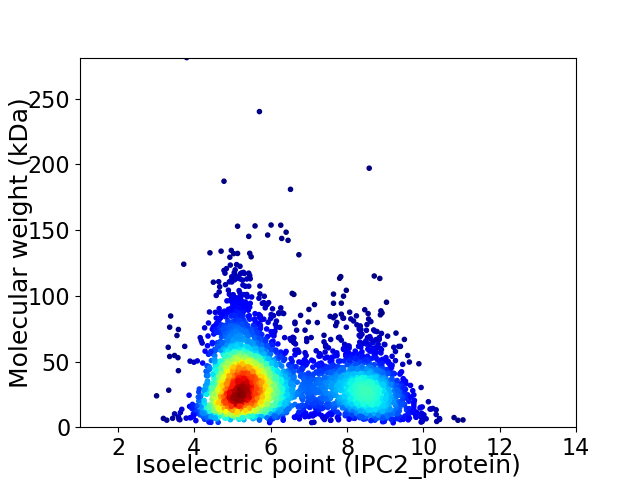
Virtual 2D-PAGE plot for 3807 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
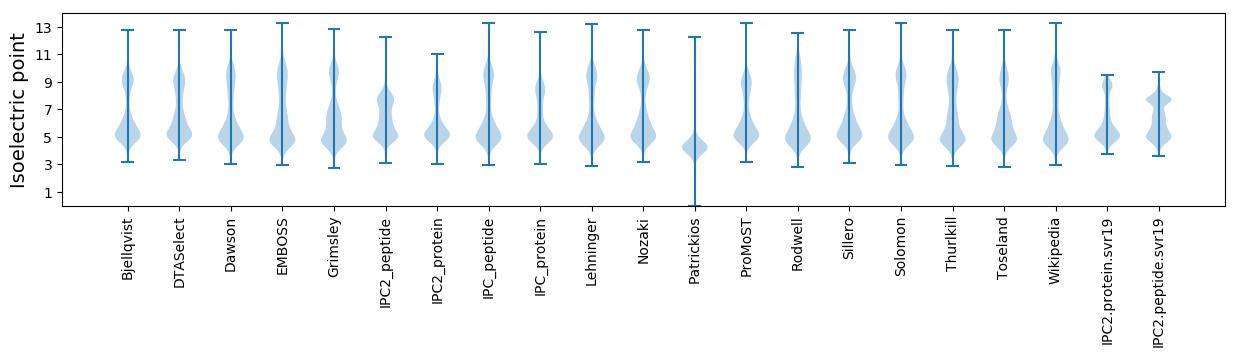
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6BZY2|C6BZY2_DESAD Extracellular solute-binding protein family 3 OS=Desulfovibrio salexigens (strain ATCC 14822 / DSM 2638 / NCIMB 8403 / VKM B-1763) OX=526222 GN=Desal_2801 PE=3 SV=1
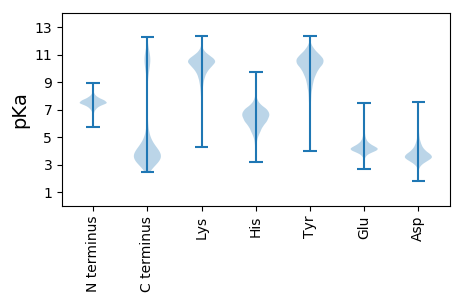
MM1 pKa = 7.55SLTNTLSIGQRR12 pKa = 11.84ALANAQVSINTTSNNIANSEE32 pKa = 4.23TEE34 pKa = 4.34GYY36 pKa = 9.81QRR38 pKa = 11.84ADD40 pKa = 2.9AVYY43 pKa = 10.67DD44 pKa = 3.4SLGNINSYY52 pKa = 11.05GNSLGTGADD61 pKa = 2.85IVAIRR66 pKa = 11.84ANWDD70 pKa = 3.35RR71 pKa = 11.84FIEE74 pKa = 4.17KK75 pKa = 10.13QFLIALGSLSCSEE88 pKa = 4.19AQLGYY93 pKa = 10.17LSQMDD98 pKa = 3.87SVFNQSEE105 pKa = 4.38EE106 pKa = 3.97QGLAAAQDD114 pKa = 3.97EE115 pKa = 5.36FLSAWNGLSTYY126 pKa = 9.76PDD128 pKa = 3.28SLAEE132 pKa = 4.27RR133 pKa = 11.84EE134 pKa = 4.27DD135 pKa = 3.83LLGEE139 pKa = 4.32AEE141 pKa = 4.17SLLYY145 pKa = 10.62GLNSSYY151 pKa = 11.48SEE153 pKa = 4.17LRR155 pKa = 11.84DD156 pKa = 3.17MSASVEE162 pKa = 4.18SEE164 pKa = 3.55IVDD167 pKa = 3.66QVNTANEE174 pKa = 4.64LIDD177 pKa = 5.27SIGLLNEE184 pKa = 4.21QISANPDD191 pKa = 3.01NYY193 pKa = 10.81EE194 pKa = 3.94LVASRR199 pKa = 11.84DD200 pKa = 3.48QAIRR204 pKa = 11.84EE205 pKa = 4.1LDD207 pKa = 3.44EE208 pKa = 5.18LIGVEE213 pKa = 4.13VLSYY217 pKa = 11.3EE218 pKa = 4.42DD219 pKa = 4.74GSTKK223 pKa = 9.95IYY225 pKa = 10.61TEE227 pKa = 4.01TGHH230 pKa = 6.73PLVEE234 pKa = 4.66GEE236 pKa = 4.41EE237 pKa = 4.24THH239 pKa = 6.56HH240 pKa = 6.96LAVAYY245 pKa = 10.36DD246 pKa = 4.39DD247 pKa = 4.71DD248 pKa = 4.19TSKK251 pKa = 9.73TGLFWEE257 pKa = 4.7NGSGGLVDD265 pKa = 4.12ITPMSDD271 pKa = 2.94EE272 pKa = 4.66SGDD275 pKa = 3.77AVSGRR280 pKa = 11.84ITGGSIAGLFITRR293 pKa = 11.84DD294 pKa = 3.31EE295 pKa = 5.15HH296 pKa = 6.48IQPTLDD302 pKa = 4.06RR303 pKa = 11.84LDD305 pKa = 4.0EE306 pKa = 4.09YY307 pKa = 11.41AAALIWEE314 pKa = 4.61TNRR317 pKa = 11.84AHH319 pKa = 5.95SQGAGLEE326 pKa = 3.96PHH328 pKa = 6.33TAVEE332 pKa = 4.2GTYY335 pKa = 10.67GVEE338 pKa = 4.15DD339 pKa = 3.4QTAALSDD346 pKa = 3.66SGLDD350 pKa = 3.47FEE352 pKa = 6.28DD353 pKa = 5.9RR354 pKa = 11.84ITSGEE359 pKa = 4.18FTIHH363 pKa = 6.68TYY365 pKa = 10.8DD366 pKa = 4.0ADD368 pKa = 4.24GNPEE372 pKa = 3.69ASITISIDD380 pKa = 3.13PSTDD384 pKa = 3.15SLDD387 pKa = 5.4DD388 pKa = 3.24IVSNINASLGGSLTASVNADD408 pKa = 3.18GEE410 pKa = 4.38LAIEE414 pKa = 4.18AVGDD418 pKa = 3.65SSFEE422 pKa = 4.0FGEE425 pKa = 4.45DD426 pKa = 3.02SSGFLAAAGINTFFEE441 pKa = 5.0GSSAGDD447 pKa = 3.15IAVNNYY453 pKa = 8.3VASNPSHH460 pKa = 6.81LNAGEE465 pKa = 4.13VGSDD469 pKa = 3.28GTVASGSNSTAQSINDD485 pKa = 3.86LLSRR489 pKa = 11.84DD490 pKa = 3.57VSIGEE495 pKa = 4.25GANATSATLTEE506 pKa = 4.31YY507 pKa = 11.12LSAIVSDD514 pKa = 3.87VGAAASTMEE523 pKa = 4.16TRR525 pKa = 11.84VTCDD529 pKa = 2.58TAAAQIYY536 pKa = 10.31AEE538 pKa = 4.16QQEE541 pKa = 4.89SVSGVNVDD549 pKa = 3.78EE550 pKa = 4.55EE551 pKa = 4.69LVNLTRR557 pKa = 11.84HH558 pKa = 4.25QQQYY562 pKa = 9.48EE563 pKa = 4.1AACQIISVTRR573 pKa = 11.84DD574 pKa = 3.12MIDD577 pKa = 3.05TVLGIVV583 pKa = 3.54
MM1 pKa = 7.55SLTNTLSIGQRR12 pKa = 11.84ALANAQVSINTTSNNIANSEE32 pKa = 4.23TEE34 pKa = 4.34GYY36 pKa = 9.81QRR38 pKa = 11.84ADD40 pKa = 2.9AVYY43 pKa = 10.67DD44 pKa = 3.4SLGNINSYY52 pKa = 11.05GNSLGTGADD61 pKa = 2.85IVAIRR66 pKa = 11.84ANWDD70 pKa = 3.35RR71 pKa = 11.84FIEE74 pKa = 4.17KK75 pKa = 10.13QFLIALGSLSCSEE88 pKa = 4.19AQLGYY93 pKa = 10.17LSQMDD98 pKa = 3.87SVFNQSEE105 pKa = 4.38EE106 pKa = 3.97QGLAAAQDD114 pKa = 3.97EE115 pKa = 5.36FLSAWNGLSTYY126 pKa = 9.76PDD128 pKa = 3.28SLAEE132 pKa = 4.27RR133 pKa = 11.84EE134 pKa = 4.27DD135 pKa = 3.83LLGEE139 pKa = 4.32AEE141 pKa = 4.17SLLYY145 pKa = 10.62GLNSSYY151 pKa = 11.48SEE153 pKa = 4.17LRR155 pKa = 11.84DD156 pKa = 3.17MSASVEE162 pKa = 4.18SEE164 pKa = 3.55IVDD167 pKa = 3.66QVNTANEE174 pKa = 4.64LIDD177 pKa = 5.27SIGLLNEE184 pKa = 4.21QISANPDD191 pKa = 3.01NYY193 pKa = 10.81EE194 pKa = 3.94LVASRR199 pKa = 11.84DD200 pKa = 3.48QAIRR204 pKa = 11.84EE205 pKa = 4.1LDD207 pKa = 3.44EE208 pKa = 5.18LIGVEE213 pKa = 4.13VLSYY217 pKa = 11.3EE218 pKa = 4.42DD219 pKa = 4.74GSTKK223 pKa = 9.95IYY225 pKa = 10.61TEE227 pKa = 4.01TGHH230 pKa = 6.73PLVEE234 pKa = 4.66GEE236 pKa = 4.41EE237 pKa = 4.24THH239 pKa = 6.56HH240 pKa = 6.96LAVAYY245 pKa = 10.36DD246 pKa = 4.39DD247 pKa = 4.71DD248 pKa = 4.19TSKK251 pKa = 9.73TGLFWEE257 pKa = 4.7NGSGGLVDD265 pKa = 4.12ITPMSDD271 pKa = 2.94EE272 pKa = 4.66SGDD275 pKa = 3.77AVSGRR280 pKa = 11.84ITGGSIAGLFITRR293 pKa = 11.84DD294 pKa = 3.31EE295 pKa = 5.15HH296 pKa = 6.48IQPTLDD302 pKa = 4.06RR303 pKa = 11.84LDD305 pKa = 4.0EE306 pKa = 4.09YY307 pKa = 11.41AAALIWEE314 pKa = 4.61TNRR317 pKa = 11.84AHH319 pKa = 5.95SQGAGLEE326 pKa = 3.96PHH328 pKa = 6.33TAVEE332 pKa = 4.2GTYY335 pKa = 10.67GVEE338 pKa = 4.15DD339 pKa = 3.4QTAALSDD346 pKa = 3.66SGLDD350 pKa = 3.47FEE352 pKa = 6.28DD353 pKa = 5.9RR354 pKa = 11.84ITSGEE359 pKa = 4.18FTIHH363 pKa = 6.68TYY365 pKa = 10.8DD366 pKa = 4.0ADD368 pKa = 4.24GNPEE372 pKa = 3.69ASITISIDD380 pKa = 3.13PSTDD384 pKa = 3.15SLDD387 pKa = 5.4DD388 pKa = 3.24IVSNINASLGGSLTASVNADD408 pKa = 3.18GEE410 pKa = 4.38LAIEE414 pKa = 4.18AVGDD418 pKa = 3.65SSFEE422 pKa = 4.0FGEE425 pKa = 4.45DD426 pKa = 3.02SSGFLAAAGINTFFEE441 pKa = 5.0GSSAGDD447 pKa = 3.15IAVNNYY453 pKa = 8.3VASNPSHH460 pKa = 6.81LNAGEE465 pKa = 4.13VGSDD469 pKa = 3.28GTVASGSNSTAQSINDD485 pKa = 3.86LLSRR489 pKa = 11.84DD490 pKa = 3.57VSIGEE495 pKa = 4.25GANATSATLTEE506 pKa = 4.31YY507 pKa = 11.12LSAIVSDD514 pKa = 3.87VGAAASTMEE523 pKa = 4.16TRR525 pKa = 11.84VTCDD529 pKa = 2.58TAAAQIYY536 pKa = 10.31AEE538 pKa = 4.16QQEE541 pKa = 4.89SVSGVNVDD549 pKa = 3.78EE550 pKa = 4.55EE551 pKa = 4.69LVNLTRR557 pKa = 11.84HH558 pKa = 4.25QQQYY562 pKa = 9.48EE563 pKa = 4.1AACQIISVTRR573 pKa = 11.84DD574 pKa = 3.12MIDD577 pKa = 3.05TVLGIVV583 pKa = 3.54
Molecular weight: 61.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6BXY8|C6BXY8_DESAD Metallo-beta-lactamase family protein OS=Desulfovibrio salexigens (strain ATCC 14822 / DSM 2638 / NCIMB 8403 / VKM B-1763) OX=526222 GN=Desal_2462 PE=4 SV=1
MM1 pKa = 6.98STLARR6 pKa = 11.84LALFGAIGTLLFLPLVVKK24 pKa = 10.41RR25 pKa = 11.84KK26 pKa = 8.14HH27 pKa = 4.72RR28 pKa = 11.84HH29 pKa = 3.79RR30 pKa = 11.84HH31 pKa = 3.83RR32 pKa = 11.84HH33 pKa = 4.51GSPVLVRR40 pKa = 11.84AGGSGGKK47 pKa = 9.6KK48 pKa = 9.23GRR50 pKa = 11.84KK51 pKa = 8.31
MM1 pKa = 6.98STLARR6 pKa = 11.84LALFGAIGTLLFLPLVVKK24 pKa = 10.41RR25 pKa = 11.84KK26 pKa = 8.14HH27 pKa = 4.72RR28 pKa = 11.84HH29 pKa = 3.79RR30 pKa = 11.84HH31 pKa = 3.83RR32 pKa = 11.84HH33 pKa = 4.51GSPVLVRR40 pKa = 11.84AGGSGGKK47 pKa = 9.6KK48 pKa = 9.23GRR50 pKa = 11.84KK51 pKa = 8.31
Molecular weight: 5.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1264146 |
31 |
2646 |
332.1 |
36.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.24 ± 0.04 | 1.335 ± 0.017 |
5.646 ± 0.033 | 7.084 ± 0.037 |
4.367 ± 0.025 | 7.518 ± 0.037 |
1.858 ± 0.017 | 6.598 ± 0.033 |
6.271 ± 0.036 | 9.818 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.967 ± 0.02 | 3.907 ± 0.026 |
4.159 ± 0.029 | 3.129 ± 0.021 |
4.81 ± 0.033 | 6.396 ± 0.032 |
4.938 ± 0.031 | 6.952 ± 0.033 |
1.062 ± 0.016 | 2.942 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
