
Varicella-zoster virus (strain Dumas) (HHV-3) (Human herpesvirus 3)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Varicellovirus; Human alphaherpesvirus 3
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
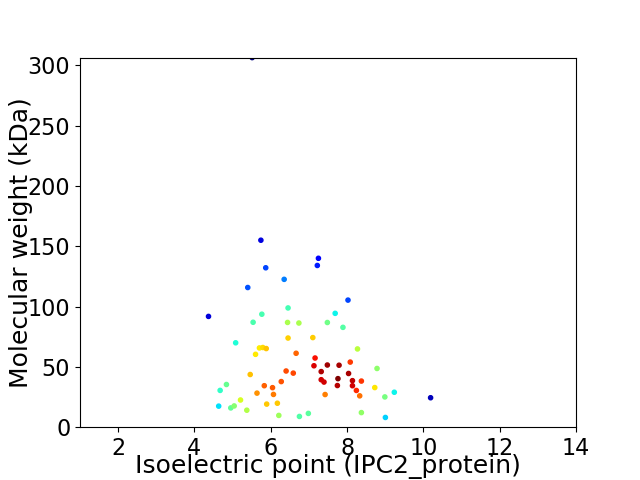
Virtual 2D-PAGE plot for 70 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P09264|TEG1_VZVD Tegument protein UL46 homolog OS=Varicella-zoster virus (strain Dumas) OX=10338 GN=ORF12 PE=3 SV=1
MM1 pKa = 7.68QSGHH5 pKa = 4.86YY6 pKa = 9.77NRR8 pKa = 11.84RR9 pKa = 11.84QSRR12 pKa = 11.84RR13 pKa = 11.84QRR15 pKa = 11.84ISSNTTDD22 pKa = 3.7SPRR25 pKa = 11.84HH26 pKa = 4.12THH28 pKa = 4.18GTRR31 pKa = 11.84YY32 pKa = 10.06RR33 pKa = 11.84STNWYY38 pKa = 5.81THH40 pKa = 6.65PPQILSNSEE49 pKa = 3.81TLVAVQEE56 pKa = 4.25LLNSEE61 pKa = 4.36MDD63 pKa = 3.34QDD65 pKa = 4.13SSSDD69 pKa = 3.47ASDD72 pKa = 4.6DD73 pKa = 3.89FPGYY77 pKa = 10.4ALHH80 pKa = 6.96HH81 pKa = 5.29STYY84 pKa = 10.77NGSEE88 pKa = 4.21QNTSTSRR95 pKa = 11.84HH96 pKa = 4.88EE97 pKa = 3.85NRR99 pKa = 11.84IFKK102 pKa = 9.06LTEE105 pKa = 3.57RR106 pKa = 11.84EE107 pKa = 4.07ANEE110 pKa = 4.39EE111 pKa = 3.9ININTDD117 pKa = 3.47AIDD120 pKa = 4.23DD121 pKa = 3.88EE122 pKa = 5.47GEE124 pKa = 4.14AEE126 pKa = 4.04EE127 pKa = 5.88GEE129 pKa = 4.29AEE131 pKa = 4.11EE132 pKa = 5.78DD133 pKa = 4.16AIDD136 pKa = 4.58DD137 pKa = 3.75EE138 pKa = 5.54GEE140 pKa = 4.14AEE142 pKa = 4.04EE143 pKa = 5.88GEE145 pKa = 4.29AEE147 pKa = 4.11EE148 pKa = 5.78DD149 pKa = 4.16AIDD152 pKa = 4.58DD153 pKa = 3.75EE154 pKa = 5.54GEE156 pKa = 4.14AEE158 pKa = 4.04EE159 pKa = 5.88GEE161 pKa = 4.29AEE163 pKa = 4.11EE164 pKa = 5.78DD165 pKa = 4.16AIDD168 pKa = 4.58DD169 pKa = 3.75EE170 pKa = 5.54GEE172 pKa = 4.14AEE174 pKa = 4.03EE175 pKa = 5.47GEE177 pKa = 4.31AEE179 pKa = 4.05EE180 pKa = 5.49GEE182 pKa = 4.31AEE184 pKa = 4.04EE185 pKa = 5.9GEE187 pKa = 4.29AEE189 pKa = 4.11EE190 pKa = 5.78DD191 pKa = 4.16AIDD194 pKa = 4.58DD195 pKa = 3.87EE196 pKa = 5.74GEE198 pKa = 4.18AEE200 pKa = 3.83EE201 pKa = 5.46DD202 pKa = 3.5AAEE205 pKa = 4.25EE206 pKa = 4.65DD207 pKa = 4.49AIDD210 pKa = 4.53DD211 pKa = 3.87EE212 pKa = 5.67GEE214 pKa = 4.0AEE216 pKa = 3.85EE217 pKa = 5.97DD218 pKa = 3.95YY219 pKa = 11.25FSVSQVCSRR228 pKa = 11.84DD229 pKa = 3.05ADD231 pKa = 3.64EE232 pKa = 6.18VYY234 pKa = 8.95FTLDD238 pKa = 3.33PEE240 pKa = 4.33ISYY243 pKa = 8.34STDD246 pKa = 2.73LRR248 pKa = 11.84IAKK251 pKa = 9.77VMEE254 pKa = 4.16PAVSKK259 pKa = 10.2EE260 pKa = 3.74LNVSKK265 pKa = 10.7RR266 pKa = 11.84CVEE269 pKa = 3.96PVTLTGSMLAHH280 pKa = 6.57NGFDD284 pKa = 3.63EE285 pKa = 4.17SWFAMRR291 pKa = 11.84EE292 pKa = 4.19CTRR295 pKa = 11.84RR296 pKa = 11.84EE297 pKa = 4.12YY298 pKa = 9.93ITVQGLYY305 pKa = 10.79DD306 pKa = 5.17PIHH309 pKa = 6.65LRR311 pKa = 11.84YY312 pKa = 10.02QFDD315 pKa = 4.0TSRR318 pKa = 11.84MTPPQILRR326 pKa = 11.84TIPALPNMTLGEE338 pKa = 4.43LLLIFPIEE346 pKa = 4.47FMAQPISIEE355 pKa = 4.23RR356 pKa = 11.84ILVEE360 pKa = 5.63DD361 pKa = 3.56VFLDD365 pKa = 3.68RR366 pKa = 11.84RR367 pKa = 11.84ASSKK371 pKa = 5.67THH373 pKa = 6.45KK374 pKa = 10.58YY375 pKa = 8.36GPRR378 pKa = 11.84WNSVYY383 pKa = 10.9ALPYY387 pKa = 9.82NAGKK391 pKa = 9.69MYY393 pKa = 10.06VQHH396 pKa = 6.92IPGFYY401 pKa = 10.01DD402 pKa = 3.13VSLRR406 pKa = 11.84AVGQGTAIWHH416 pKa = 5.64HH417 pKa = 6.6MILSTAACAISNRR430 pKa = 11.84ISHH433 pKa = 6.71GDD435 pKa = 3.3GLGFLLDD442 pKa = 3.43AAIRR446 pKa = 11.84ISANCIFLGRR456 pKa = 11.84NDD458 pKa = 3.56NFGVGDD464 pKa = 3.98PCWLEE469 pKa = 3.78DD470 pKa = 4.1HH471 pKa = 6.89LAGLPRR477 pKa = 11.84EE478 pKa = 4.31AVPDD482 pKa = 3.81VLQVTQLVLPNRR494 pKa = 11.84GPTVAIMRR502 pKa = 11.84GFFGALAYY510 pKa = 9.6WPEE513 pKa = 3.68LRR515 pKa = 11.84IAISEE520 pKa = 4.18PSTSLVRR527 pKa = 11.84YY528 pKa = 7.33ATGHH532 pKa = 5.77MEE534 pKa = 3.77LAEE537 pKa = 4.01WFLFSRR543 pKa = 11.84THH545 pKa = 5.1SLKK548 pKa = 10.65PQFTPTEE555 pKa = 4.19RR556 pKa = 11.84EE557 pKa = 3.74MLASFFTLYY566 pKa = 9.1VTLGGGMLNWICRR579 pKa = 11.84ATAMYY584 pKa = 10.16LAAPYY589 pKa = 9.47HH590 pKa = 6.23SRR592 pKa = 11.84SAYY595 pKa = 9.45IAVCEE600 pKa = 4.14SLPYY604 pKa = 10.67YY605 pKa = 8.97YY606 pKa = 10.01IPVNSDD612 pKa = 3.64LLCDD616 pKa = 4.78LEE618 pKa = 4.37VLLLGEE624 pKa = 4.17VDD626 pKa = 4.74LPTVCEE632 pKa = 4.3SYY634 pKa = 10.13ATIAHH639 pKa = 6.82EE640 pKa = 4.12LTGYY644 pKa = 9.72EE645 pKa = 4.23AVRR648 pKa = 11.84TAATNFMIEE657 pKa = 3.97FADD660 pKa = 4.32CYY662 pKa = 10.81KK663 pKa = 10.78EE664 pKa = 4.58SEE666 pKa = 4.19TDD668 pKa = 3.67LMVSAYY674 pKa = 10.36LGAVLLLQRR683 pKa = 11.84VLGHH687 pKa = 7.09ANLLLLLLSGAALYY701 pKa = 9.5GGCSIYY707 pKa = 10.23IPRR710 pKa = 11.84GILDD714 pKa = 4.32AYY716 pKa = 8.47NTLMLAASPLYY727 pKa = 10.44AHH729 pKa = 6.1QTLTSFWKK737 pKa = 10.68DD738 pKa = 2.66RR739 pKa = 11.84DD740 pKa = 3.84DD741 pKa = 5.0AMQTLGIRR749 pKa = 11.84PTTDD753 pKa = 3.01VLPKK757 pKa = 9.02EE758 pKa = 4.14QDD760 pKa = 3.58RR761 pKa = 11.84IVQASPIEE769 pKa = 4.0MNFRR773 pKa = 11.84FVGLEE778 pKa = 3.7TIYY781 pKa = 10.5PRR783 pKa = 11.84EE784 pKa = 3.9QPIPSVDD791 pKa = 3.41LAEE794 pKa = 5.39NLMQYY799 pKa = 10.47RR800 pKa = 11.84NEE802 pKa = 3.92ILGLDD807 pKa = 3.64WKK809 pKa = 10.61SVAMHH814 pKa = 7.3LLRR817 pKa = 11.84KK818 pKa = 9.19YY819 pKa = 10.73
MM1 pKa = 7.68QSGHH5 pKa = 4.86YY6 pKa = 9.77NRR8 pKa = 11.84RR9 pKa = 11.84QSRR12 pKa = 11.84RR13 pKa = 11.84QRR15 pKa = 11.84ISSNTTDD22 pKa = 3.7SPRR25 pKa = 11.84HH26 pKa = 4.12THH28 pKa = 4.18GTRR31 pKa = 11.84YY32 pKa = 10.06RR33 pKa = 11.84STNWYY38 pKa = 5.81THH40 pKa = 6.65PPQILSNSEE49 pKa = 3.81TLVAVQEE56 pKa = 4.25LLNSEE61 pKa = 4.36MDD63 pKa = 3.34QDD65 pKa = 4.13SSSDD69 pKa = 3.47ASDD72 pKa = 4.6DD73 pKa = 3.89FPGYY77 pKa = 10.4ALHH80 pKa = 6.96HH81 pKa = 5.29STYY84 pKa = 10.77NGSEE88 pKa = 4.21QNTSTSRR95 pKa = 11.84HH96 pKa = 4.88EE97 pKa = 3.85NRR99 pKa = 11.84IFKK102 pKa = 9.06LTEE105 pKa = 3.57RR106 pKa = 11.84EE107 pKa = 4.07ANEE110 pKa = 4.39EE111 pKa = 3.9ININTDD117 pKa = 3.47AIDD120 pKa = 4.23DD121 pKa = 3.88EE122 pKa = 5.47GEE124 pKa = 4.14AEE126 pKa = 4.04EE127 pKa = 5.88GEE129 pKa = 4.29AEE131 pKa = 4.11EE132 pKa = 5.78DD133 pKa = 4.16AIDD136 pKa = 4.58DD137 pKa = 3.75EE138 pKa = 5.54GEE140 pKa = 4.14AEE142 pKa = 4.04EE143 pKa = 5.88GEE145 pKa = 4.29AEE147 pKa = 4.11EE148 pKa = 5.78DD149 pKa = 4.16AIDD152 pKa = 4.58DD153 pKa = 3.75EE154 pKa = 5.54GEE156 pKa = 4.14AEE158 pKa = 4.04EE159 pKa = 5.88GEE161 pKa = 4.29AEE163 pKa = 4.11EE164 pKa = 5.78DD165 pKa = 4.16AIDD168 pKa = 4.58DD169 pKa = 3.75EE170 pKa = 5.54GEE172 pKa = 4.14AEE174 pKa = 4.03EE175 pKa = 5.47GEE177 pKa = 4.31AEE179 pKa = 4.05EE180 pKa = 5.49GEE182 pKa = 4.31AEE184 pKa = 4.04EE185 pKa = 5.9GEE187 pKa = 4.29AEE189 pKa = 4.11EE190 pKa = 5.78DD191 pKa = 4.16AIDD194 pKa = 4.58DD195 pKa = 3.87EE196 pKa = 5.74GEE198 pKa = 4.18AEE200 pKa = 3.83EE201 pKa = 5.46DD202 pKa = 3.5AAEE205 pKa = 4.25EE206 pKa = 4.65DD207 pKa = 4.49AIDD210 pKa = 4.53DD211 pKa = 3.87EE212 pKa = 5.67GEE214 pKa = 4.0AEE216 pKa = 3.85EE217 pKa = 5.97DD218 pKa = 3.95YY219 pKa = 11.25FSVSQVCSRR228 pKa = 11.84DD229 pKa = 3.05ADD231 pKa = 3.64EE232 pKa = 6.18VYY234 pKa = 8.95FTLDD238 pKa = 3.33PEE240 pKa = 4.33ISYY243 pKa = 8.34STDD246 pKa = 2.73LRR248 pKa = 11.84IAKK251 pKa = 9.77VMEE254 pKa = 4.16PAVSKK259 pKa = 10.2EE260 pKa = 3.74LNVSKK265 pKa = 10.7RR266 pKa = 11.84CVEE269 pKa = 3.96PVTLTGSMLAHH280 pKa = 6.57NGFDD284 pKa = 3.63EE285 pKa = 4.17SWFAMRR291 pKa = 11.84EE292 pKa = 4.19CTRR295 pKa = 11.84RR296 pKa = 11.84EE297 pKa = 4.12YY298 pKa = 9.93ITVQGLYY305 pKa = 10.79DD306 pKa = 5.17PIHH309 pKa = 6.65LRR311 pKa = 11.84YY312 pKa = 10.02QFDD315 pKa = 4.0TSRR318 pKa = 11.84MTPPQILRR326 pKa = 11.84TIPALPNMTLGEE338 pKa = 4.43LLLIFPIEE346 pKa = 4.47FMAQPISIEE355 pKa = 4.23RR356 pKa = 11.84ILVEE360 pKa = 5.63DD361 pKa = 3.56VFLDD365 pKa = 3.68RR366 pKa = 11.84RR367 pKa = 11.84ASSKK371 pKa = 5.67THH373 pKa = 6.45KK374 pKa = 10.58YY375 pKa = 8.36GPRR378 pKa = 11.84WNSVYY383 pKa = 10.9ALPYY387 pKa = 9.82NAGKK391 pKa = 9.69MYY393 pKa = 10.06VQHH396 pKa = 6.92IPGFYY401 pKa = 10.01DD402 pKa = 3.13VSLRR406 pKa = 11.84AVGQGTAIWHH416 pKa = 5.64HH417 pKa = 6.6MILSTAACAISNRR430 pKa = 11.84ISHH433 pKa = 6.71GDD435 pKa = 3.3GLGFLLDD442 pKa = 3.43AAIRR446 pKa = 11.84ISANCIFLGRR456 pKa = 11.84NDD458 pKa = 3.56NFGVGDD464 pKa = 3.98PCWLEE469 pKa = 3.78DD470 pKa = 4.1HH471 pKa = 6.89LAGLPRR477 pKa = 11.84EE478 pKa = 4.31AVPDD482 pKa = 3.81VLQVTQLVLPNRR494 pKa = 11.84GPTVAIMRR502 pKa = 11.84GFFGALAYY510 pKa = 9.6WPEE513 pKa = 3.68LRR515 pKa = 11.84IAISEE520 pKa = 4.18PSTSLVRR527 pKa = 11.84YY528 pKa = 7.33ATGHH532 pKa = 5.77MEE534 pKa = 3.77LAEE537 pKa = 4.01WFLFSRR543 pKa = 11.84THH545 pKa = 5.1SLKK548 pKa = 10.65PQFTPTEE555 pKa = 4.19RR556 pKa = 11.84EE557 pKa = 3.74MLASFFTLYY566 pKa = 9.1VTLGGGMLNWICRR579 pKa = 11.84ATAMYY584 pKa = 10.16LAAPYY589 pKa = 9.47HH590 pKa = 6.23SRR592 pKa = 11.84SAYY595 pKa = 9.45IAVCEE600 pKa = 4.14SLPYY604 pKa = 10.67YY605 pKa = 8.97YY606 pKa = 10.01IPVNSDD612 pKa = 3.64LLCDD616 pKa = 4.78LEE618 pKa = 4.37VLLLGEE624 pKa = 4.17VDD626 pKa = 4.74LPTVCEE632 pKa = 4.3SYY634 pKa = 10.13ATIAHH639 pKa = 6.82EE640 pKa = 4.12LTGYY644 pKa = 9.72EE645 pKa = 4.23AVRR648 pKa = 11.84TAATNFMIEE657 pKa = 3.97FADD660 pKa = 4.32CYY662 pKa = 10.81KK663 pKa = 10.78EE664 pKa = 4.58SEE666 pKa = 4.19TDD668 pKa = 3.67LMVSAYY674 pKa = 10.36LGAVLLLQRR683 pKa = 11.84VLGHH687 pKa = 7.09ANLLLLLLSGAALYY701 pKa = 9.5GGCSIYY707 pKa = 10.23IPRR710 pKa = 11.84GILDD714 pKa = 4.32AYY716 pKa = 8.47NTLMLAASPLYY727 pKa = 10.44AHH729 pKa = 6.1QTLTSFWKK737 pKa = 10.68DD738 pKa = 2.66RR739 pKa = 11.84DD740 pKa = 3.84DD741 pKa = 5.0AMQTLGIRR749 pKa = 11.84PTTDD753 pKa = 3.01VLPKK757 pKa = 9.02EE758 pKa = 4.14QDD760 pKa = 3.58RR761 pKa = 11.84IVQASPIEE769 pKa = 4.0MNFRR773 pKa = 11.84FVGLEE778 pKa = 3.7TIYY781 pKa = 10.5PRR783 pKa = 11.84EE784 pKa = 3.9QPIPSVDD791 pKa = 3.41LAEE794 pKa = 5.39NLMQYY799 pKa = 10.47RR800 pKa = 11.84NEE802 pKa = 3.92ILGLDD807 pKa = 3.64WKK809 pKa = 10.61SVAMHH814 pKa = 7.3LLRR817 pKa = 11.84KK818 pKa = 9.19YY819 pKa = 10.73
Molecular weight: 91.83 kDa
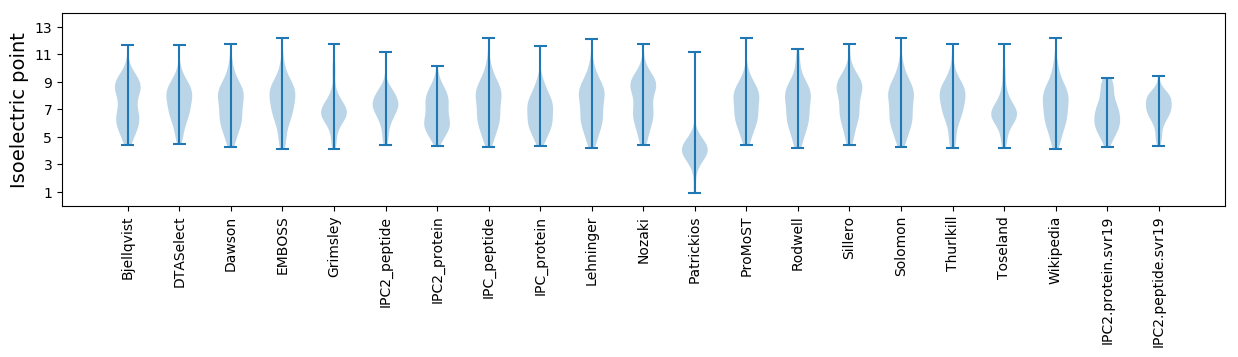
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P09289|TEG4_VZVD Tegument protein UL21 homolog OS=Varicella-zoster virus (strain Dumas) OX=10338 GN=ORF38 PE=3 SV=1
MM1 pKa = 6.78SASRR5 pKa = 11.84IRR7 pKa = 11.84AKK9 pKa = 10.37CFRR12 pKa = 11.84LGQRR16 pKa = 11.84CHH18 pKa = 5.15TRR20 pKa = 11.84FYY22 pKa = 11.56DD23 pKa = 3.84VLKK26 pKa = 10.89KK27 pKa = 10.84DD28 pKa = 2.9IDD30 pKa = 3.54NVRR33 pKa = 11.84RR34 pKa = 11.84GFADD38 pKa = 3.49AFNPRR43 pKa = 11.84LAKK46 pKa = 10.44LLSPLSHH53 pKa = 7.24VDD55 pKa = 2.88VQRR58 pKa = 11.84AVRR61 pKa = 11.84ISMSFEE67 pKa = 3.86VNLGRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84PDD76 pKa = 3.31CVCIIQTEE84 pKa = 4.29SSGAGKK90 pKa = 7.25TVCFIVEE97 pKa = 4.42LKK99 pKa = 10.32SCRR102 pKa = 11.84FSANIHH108 pKa = 5.9TPTKK112 pKa = 9.62YY113 pKa = 10.09HH114 pKa = 5.37QFCEE118 pKa = 4.36GMRR121 pKa = 11.84QLRR124 pKa = 11.84DD125 pKa = 3.07TMALIKK131 pKa = 9.21EE132 pKa = 4.49TTPTGSDD139 pKa = 3.44EE140 pKa = 4.86IMVTPLLVFVSQRR153 pKa = 11.84GLNLLQVTRR162 pKa = 11.84LPPKK166 pKa = 9.79VIHH169 pKa = 6.65GNLVMLASHH178 pKa = 7.15LEE180 pKa = 4.06NVAEE184 pKa = 4.08YY185 pKa = 8.61TPPIRR190 pKa = 11.84SVRR193 pKa = 11.84EE194 pKa = 3.45RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84LCKK200 pKa = 10.2KK201 pKa = 10.03KK202 pKa = 10.17IHH204 pKa = 5.83VCSLAKK210 pKa = 10.12KK211 pKa = 9.26RR212 pKa = 11.84AKK214 pKa = 9.87SCHH217 pKa = 6.0RR218 pKa = 11.84SALTKK223 pKa = 10.46FEE225 pKa = 4.52EE226 pKa = 4.34NAACGVDD233 pKa = 3.79LPLRR237 pKa = 11.84RR238 pKa = 11.84PSLGACGGILQSITGMFSHH257 pKa = 7.15GG258 pKa = 3.37
MM1 pKa = 6.78SASRR5 pKa = 11.84IRR7 pKa = 11.84AKK9 pKa = 10.37CFRR12 pKa = 11.84LGQRR16 pKa = 11.84CHH18 pKa = 5.15TRR20 pKa = 11.84FYY22 pKa = 11.56DD23 pKa = 3.84VLKK26 pKa = 10.89KK27 pKa = 10.84DD28 pKa = 2.9IDD30 pKa = 3.54NVRR33 pKa = 11.84RR34 pKa = 11.84GFADD38 pKa = 3.49AFNPRR43 pKa = 11.84LAKK46 pKa = 10.44LLSPLSHH53 pKa = 7.24VDD55 pKa = 2.88VQRR58 pKa = 11.84AVRR61 pKa = 11.84ISMSFEE67 pKa = 3.86VNLGRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84PDD76 pKa = 3.31CVCIIQTEE84 pKa = 4.29SSGAGKK90 pKa = 7.25TVCFIVEE97 pKa = 4.42LKK99 pKa = 10.32SCRR102 pKa = 11.84FSANIHH108 pKa = 5.9TPTKK112 pKa = 9.62YY113 pKa = 10.09HH114 pKa = 5.37QFCEE118 pKa = 4.36GMRR121 pKa = 11.84QLRR124 pKa = 11.84DD125 pKa = 3.07TMALIKK131 pKa = 9.21EE132 pKa = 4.49TTPTGSDD139 pKa = 3.44EE140 pKa = 4.86IMVTPLLVFVSQRR153 pKa = 11.84GLNLLQVTRR162 pKa = 11.84LPPKK166 pKa = 9.79VIHH169 pKa = 6.65GNLVMLASHH178 pKa = 7.15LEE180 pKa = 4.06NVAEE184 pKa = 4.08YY185 pKa = 8.61TPPIRR190 pKa = 11.84SVRR193 pKa = 11.84EE194 pKa = 3.45RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84LCKK200 pKa = 10.2KK201 pKa = 10.03KK202 pKa = 10.17IHH204 pKa = 5.83VCSLAKK210 pKa = 10.12KK211 pKa = 9.26RR212 pKa = 11.84AKK214 pKa = 9.87SCHH217 pKa = 6.0RR218 pKa = 11.84SALTKK223 pKa = 10.46FEE225 pKa = 4.52EE226 pKa = 4.34NAACGVDD233 pKa = 3.79LPLRR237 pKa = 11.84RR238 pKa = 11.84PSLGACGGILQSITGMFSHH257 pKa = 7.15GG258 pKa = 3.37
Molecular weight: 28.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
36084 |
71 |
2763 |
515.5 |
57.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.43 ± 0.196 | 2.148 ± 0.123 |
5.254 ± 0.155 | 5.091 ± 0.177 |
4.157 ± 0.167 | 6.03 ± 0.211 |
2.638 ± 0.101 | 5.695 ± 0.206 |
3.564 ± 0.154 | 9.708 ± 0.209 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.128 ± 0.092 | 4.199 ± 0.128 |
5.886 ± 0.251 | 3.553 ± 0.115 |
6.391 ± 0.211 | 7.341 ± 0.169 |
7.422 ± 0.204 | 6.862 ± 0.164 |
1.059 ± 0.063 | 3.445 ± 0.132 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
