
Klebsiella phage ST437-OXA245phi4.1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Felsduovirus; Klebsiella virus ST437OXA245phi4-1
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
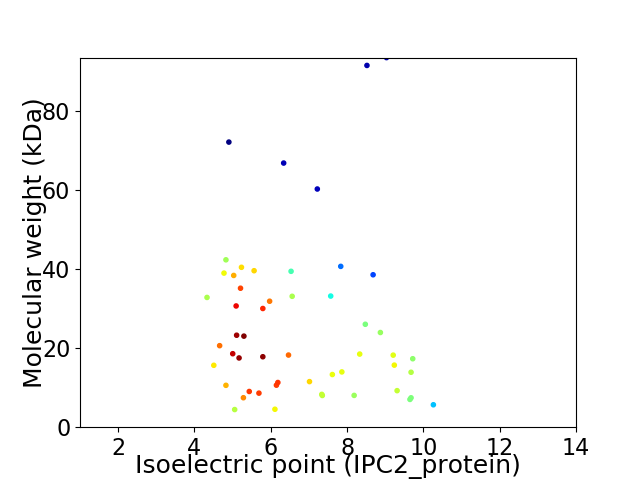
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A482IMP6|A0A482IMP6_9CAUD Tail protein OS=Klebsiella phage ST437-OXA245phi4.1 OX=2510486 PE=4 SV=1
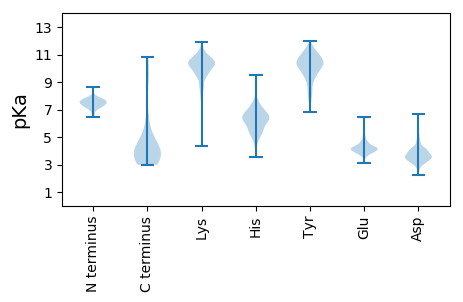
MM1 pKa = 7.42AVIDD5 pKa = 4.87LSQLPPPQIVDD16 pKa = 3.52EE17 pKa = 4.79PDD19 pKa = 3.3FEE21 pKa = 4.61TLLTEE26 pKa = 4.69RR27 pKa = 11.84KK28 pKa = 9.86AEE30 pKa = 4.22FVALYY35 pKa = 9.26PAEE38 pKa = 4.11EE39 pKa = 4.24QEE41 pKa = 4.27AVARR45 pKa = 11.84TLTLEE50 pKa = 3.98SEE52 pKa = 4.93PIVKK56 pKa = 8.7TLQEE60 pKa = 3.77NVYY63 pKa = 10.59RR64 pKa = 11.84EE65 pKa = 3.9LLLRR69 pKa = 11.84QRR71 pKa = 11.84INEE74 pKa = 3.94AARR77 pKa = 11.84AVMVAYY83 pKa = 10.37SGSDD87 pKa = 3.34DD88 pKa = 5.21LDD90 pKa = 3.54NLGANNNVQRR100 pKa = 11.84RR101 pKa = 11.84VITPADD107 pKa = 3.79DD108 pKa = 3.51TTTPPTEE115 pKa = 4.3AEE117 pKa = 4.23MEE119 pKa = 3.94SDD121 pKa = 2.65ADD123 pKa = 3.49YY124 pKa = 10.68RR125 pKa = 11.84QRR127 pKa = 11.84IPAAFEE133 pKa = 4.07GMSVAGPVGAYY144 pKa = 8.33EE145 pKa = 4.06FHH147 pKa = 7.02ALSADD152 pKa = 3.18GRR154 pKa = 11.84VADD157 pKa = 5.22ASAFSPAPAEE167 pKa = 4.16VVVTILARR175 pKa = 11.84DD176 pKa = 3.81GDD178 pKa = 4.25GTAPDD183 pKa = 4.95DD184 pKa = 3.91LLQVVGVALNDD195 pKa = 3.39EE196 pKa = 4.53AVRR199 pKa = 11.84PVADD203 pKa = 3.34RR204 pKa = 11.84VSVRR208 pKa = 11.84SAEE211 pKa = 3.96IVRR214 pKa = 11.84YY215 pKa = 9.67EE216 pKa = 3.88IDD218 pKa = 3.18AVLYY222 pKa = 8.8VYY224 pKa = 9.63PGPAKK229 pKa = 10.45EE230 pKa = 4.92PILAAAKK237 pKa = 9.51EE238 pKa = 4.01QGTAYY243 pKa = 9.86INEE246 pKa = 3.94QRR248 pKa = 11.84RR249 pKa = 11.84LGRR252 pKa = 11.84DD253 pKa = 3.05VRR255 pKa = 11.84LSAIYY260 pKa = 9.96AALHH264 pKa = 4.63VQGVQRR270 pKa = 11.84VEE272 pKa = 4.02LMQPLADD279 pKa = 3.66MVIDD283 pKa = 3.8KK284 pKa = 8.59TQASYY289 pKa = 9.0CTDD292 pKa = 3.96FKK294 pKa = 11.81AEE296 pKa = 3.95IGGSDD301 pKa = 3.47EE302 pKa = 4.06
MM1 pKa = 7.42AVIDD5 pKa = 4.87LSQLPPPQIVDD16 pKa = 3.52EE17 pKa = 4.79PDD19 pKa = 3.3FEE21 pKa = 4.61TLLTEE26 pKa = 4.69RR27 pKa = 11.84KK28 pKa = 9.86AEE30 pKa = 4.22FVALYY35 pKa = 9.26PAEE38 pKa = 4.11EE39 pKa = 4.24QEE41 pKa = 4.27AVARR45 pKa = 11.84TLTLEE50 pKa = 3.98SEE52 pKa = 4.93PIVKK56 pKa = 8.7TLQEE60 pKa = 3.77NVYY63 pKa = 10.59RR64 pKa = 11.84EE65 pKa = 3.9LLLRR69 pKa = 11.84QRR71 pKa = 11.84INEE74 pKa = 3.94AARR77 pKa = 11.84AVMVAYY83 pKa = 10.37SGSDD87 pKa = 3.34DD88 pKa = 5.21LDD90 pKa = 3.54NLGANNNVQRR100 pKa = 11.84RR101 pKa = 11.84VITPADD107 pKa = 3.79DD108 pKa = 3.51TTTPPTEE115 pKa = 4.3AEE117 pKa = 4.23MEE119 pKa = 3.94SDD121 pKa = 2.65ADD123 pKa = 3.49YY124 pKa = 10.68RR125 pKa = 11.84QRR127 pKa = 11.84IPAAFEE133 pKa = 4.07GMSVAGPVGAYY144 pKa = 8.33EE145 pKa = 4.06FHH147 pKa = 7.02ALSADD152 pKa = 3.18GRR154 pKa = 11.84VADD157 pKa = 5.22ASAFSPAPAEE167 pKa = 4.16VVVTILARR175 pKa = 11.84DD176 pKa = 3.81GDD178 pKa = 4.25GTAPDD183 pKa = 4.95DD184 pKa = 3.91LLQVVGVALNDD195 pKa = 3.39EE196 pKa = 4.53AVRR199 pKa = 11.84PVADD203 pKa = 3.34RR204 pKa = 11.84VSVRR208 pKa = 11.84SAEE211 pKa = 3.96IVRR214 pKa = 11.84YY215 pKa = 9.67EE216 pKa = 3.88IDD218 pKa = 3.18AVLYY222 pKa = 8.8VYY224 pKa = 9.63PGPAKK229 pKa = 10.45EE230 pKa = 4.92PILAAAKK237 pKa = 9.51EE238 pKa = 4.01QGTAYY243 pKa = 9.86INEE246 pKa = 3.94QRR248 pKa = 11.84RR249 pKa = 11.84LGRR252 pKa = 11.84DD253 pKa = 3.05VRR255 pKa = 11.84LSAIYY260 pKa = 9.96AALHH264 pKa = 4.63VQGVQRR270 pKa = 11.84VEE272 pKa = 4.02LMQPLADD279 pKa = 3.66MVIDD283 pKa = 3.8KK284 pKa = 8.59TQASYY289 pKa = 9.0CTDD292 pKa = 3.96FKK294 pKa = 11.81AEE296 pKa = 3.95IGGSDD301 pKa = 3.47EE302 pKa = 4.06
Molecular weight: 32.84 kDa
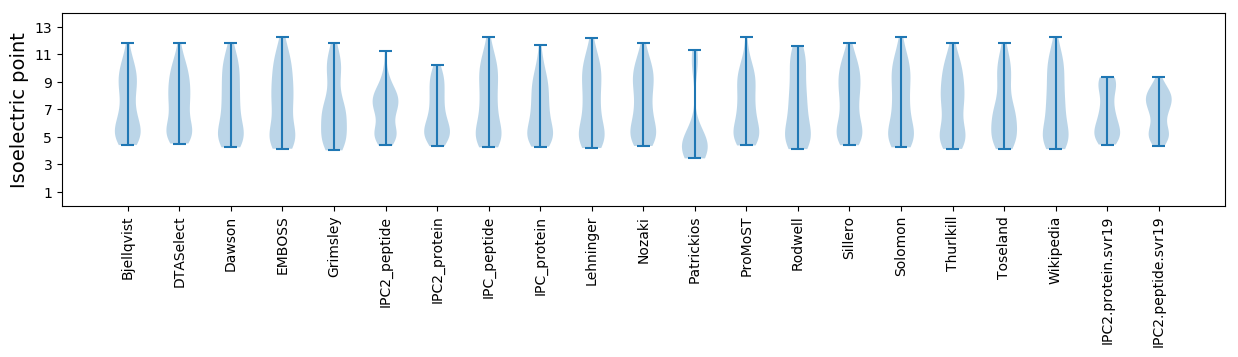
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482IM29|A0A482IM29_9CAUD Capsid scaffolding protein OS=Klebsiella phage ST437-OXA245phi4.1 OX=2510486 PE=4 SV=1
MM1 pKa = 7.95AEE3 pKa = 4.03LQEE6 pKa = 3.83VDD8 pKa = 3.12AWLEE12 pKa = 3.72ALLAGLEE19 pKa = 3.9PAARR23 pKa = 11.84KK24 pKa = 10.25RR25 pKa = 11.84MMRR28 pKa = 11.84EE29 pKa = 3.45LAQQLRR35 pKa = 11.84RR36 pKa = 11.84SQQKK40 pKa = 9.63NIRR43 pKa = 11.84MQRR46 pKa = 11.84NPDD49 pKa = 2.92GTAYY53 pKa = 10.1EE54 pKa = 4.13PRR56 pKa = 11.84RR57 pKa = 11.84VTARR61 pKa = 11.84TKK63 pKa = 8.63TGRR66 pKa = 11.84IRR68 pKa = 11.84RR69 pKa = 11.84QMFAKK74 pKa = 10.34LRR76 pKa = 11.84TAKK79 pKa = 10.07YY80 pKa = 9.37LKK82 pKa = 10.42AAASPDD88 pKa = 3.6SASVEE93 pKa = 4.08FEE95 pKa = 3.66GRR97 pKa = 11.84VQRR100 pKa = 11.84IARR103 pKa = 11.84VHH105 pKa = 6.27HH106 pKa = 5.92YY107 pKa = 10.35GLRR110 pKa = 11.84DD111 pKa = 3.17RR112 pKa = 11.84VSRR115 pKa = 11.84RR116 pKa = 11.84GPEE119 pKa = 3.66VQYY122 pKa = 10.0SQRR125 pKa = 11.84RR126 pKa = 11.84LLGINDD132 pKa = 3.72EE133 pKa = 4.51VEE135 pKa = 4.89DD136 pKa = 3.74ITRR139 pKa = 11.84EE140 pKa = 3.94TLLDD144 pKa = 4.71WIQKK148 pKa = 9.41
MM1 pKa = 7.95AEE3 pKa = 4.03LQEE6 pKa = 3.83VDD8 pKa = 3.12AWLEE12 pKa = 3.72ALLAGLEE19 pKa = 3.9PAARR23 pKa = 11.84KK24 pKa = 10.25RR25 pKa = 11.84MMRR28 pKa = 11.84EE29 pKa = 3.45LAQQLRR35 pKa = 11.84RR36 pKa = 11.84SQQKK40 pKa = 9.63NIRR43 pKa = 11.84MQRR46 pKa = 11.84NPDD49 pKa = 2.92GTAYY53 pKa = 10.1EE54 pKa = 4.13PRR56 pKa = 11.84RR57 pKa = 11.84VTARR61 pKa = 11.84TKK63 pKa = 8.63TGRR66 pKa = 11.84IRR68 pKa = 11.84RR69 pKa = 11.84QMFAKK74 pKa = 10.34LRR76 pKa = 11.84TAKK79 pKa = 10.07YY80 pKa = 9.37LKK82 pKa = 10.42AAASPDD88 pKa = 3.6SASVEE93 pKa = 4.08FEE95 pKa = 3.66GRR97 pKa = 11.84VQRR100 pKa = 11.84IARR103 pKa = 11.84VHH105 pKa = 6.27HH106 pKa = 5.92YY107 pKa = 10.35GLRR110 pKa = 11.84DD111 pKa = 3.17RR112 pKa = 11.84VSRR115 pKa = 11.84RR116 pKa = 11.84GPEE119 pKa = 3.66VQYY122 pKa = 10.0SQRR125 pKa = 11.84RR126 pKa = 11.84LLGINDD132 pKa = 3.72EE133 pKa = 4.51VEE135 pKa = 4.89DD136 pKa = 3.74ITRR139 pKa = 11.84EE140 pKa = 3.94TLLDD144 pKa = 4.71WIQKK148 pKa = 9.41
Molecular weight: 17.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12309 |
37 |
876 |
232.2 |
26.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.27 ± 0.552 | 1.105 ± 0.14 |
5.809 ± 0.249 | 6.272 ± 0.354 |
3.558 ± 0.22 | 6.556 ± 0.38 |
2.08 ± 0.195 | 5.565 ± 0.295 |
5.427 ± 0.324 | 9.538 ± 0.306 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.77 ± 0.163 | 4.29 ± 0.217 |
4.192 ± 0.236 | 4.233 ± 0.233 |
6.442 ± 0.424 | 6.207 ± 0.226 |
5.833 ± 0.333 | 6.207 ± 0.301 |
1.535 ± 0.141 | 3.112 ± 0.214 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
