
candidate division Zixibacteria bacterium RBG-1
Taxonomy: cellular organisms; Bacteria; FCB group; candidate division Zixibacteria; unclassified candidate division Zixibacteria
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
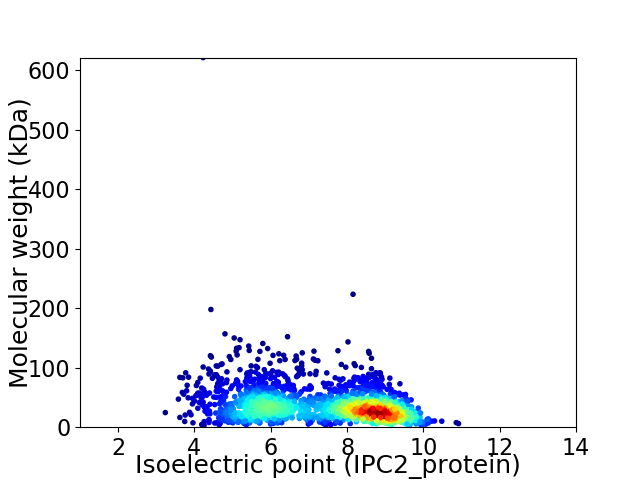
Virtual 2D-PAGE plot for 1906 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T0MRB1|T0MRB1_9BACT Uncharacterized protein OS=candidate division Zixibacteria bacterium RBG-1 OX=1379698 GN=RBG1_1C00001G1274 PE=4 SV=1
MM1 pKa = 7.42GRR3 pKa = 11.84LDD5 pKa = 3.51LSLRR9 pKa = 11.84HH10 pKa = 6.16PPSLTSGIPQGDD22 pKa = 3.53NVNLLKK28 pKa = 10.82RR29 pKa = 11.84GVKK32 pKa = 9.64KK33 pKa = 10.61LKK35 pKa = 9.87KK36 pKa = 9.33SAMGLVVILIFLLASFQIAFSQNPDD61 pKa = 3.91SIPFAPAVDD70 pKa = 3.7YY71 pKa = 11.21AVGNNPDD78 pKa = 4.92DD79 pKa = 4.39NPDD82 pKa = 4.17CIFVSDD88 pKa = 4.82LDD90 pKa = 4.04GDD92 pKa = 4.11GDD94 pKa = 3.76IDD96 pKa = 4.72LAVAMSWNDD105 pKa = 3.63SISILMNNGDD115 pKa = 4.07GTFATQISYY124 pKa = 8.77PAGSYY129 pKa = 9.09PRR131 pKa = 11.84EE132 pKa = 4.05VFGSDD137 pKa = 3.25FDD139 pKa = 5.24SDD141 pKa = 3.7GDD143 pKa = 3.65TDD145 pKa = 4.2LAIANWYY152 pKa = 8.91GNSVSILKK160 pKa = 10.62NNGDD164 pKa = 3.49GTFYY168 pKa = 11.5AKK170 pKa = 10.16VDD172 pKa = 3.66YY173 pKa = 10.46DD174 pKa = 3.72AGVAPVSVFASDD186 pKa = 4.92LDD188 pKa = 3.81FDD190 pKa = 4.8GNQDD194 pKa = 3.61FVVSKK199 pKa = 10.23QSEE202 pKa = 4.24VGVFMNKK209 pKa = 9.08GDD211 pKa = 3.57ATFEE215 pKa = 4.02PRR217 pKa = 11.84VEE219 pKa = 4.19YY220 pKa = 9.36PAGGVSVYY228 pKa = 10.53ISDD231 pKa = 4.67LDD233 pKa = 3.94GDD235 pKa = 4.33GDD237 pKa = 3.99SDD239 pKa = 4.7LAVSSTGSDD248 pKa = 3.34EE249 pKa = 5.01ISILRR254 pKa = 11.84NNGDD258 pKa = 3.22GTFFPKK264 pKa = 10.34VNYY267 pKa = 8.42LTGHH271 pKa = 5.72VPTSIFVCDD280 pKa = 3.64IDD282 pKa = 5.74GDD284 pKa = 3.99TDD286 pKa = 3.28QDD288 pKa = 3.63IIVPNFSASTISVLKK303 pKa = 10.91NNGDD307 pKa = 3.46GTFATKK313 pKa = 10.27VDD315 pKa = 3.87YY316 pKa = 8.66PTEE319 pKa = 3.97YY320 pKa = 10.67SAVKK324 pKa = 10.22IIAIDD329 pKa = 3.8LDD331 pKa = 4.02LDD333 pKa = 3.89GDD335 pKa = 3.61QDD337 pKa = 4.23IIVSHH342 pKa = 6.54EE343 pKa = 4.14NNPIISIFKK352 pKa = 10.39NNGDD356 pKa = 3.42GTFPSRR362 pKa = 11.84LNYY365 pKa = 10.23SVGAGGVTHH374 pKa = 6.71WVVAADD380 pKa = 4.02LDD382 pKa = 4.35GDD384 pKa = 4.01GDD386 pKa = 4.21YY387 pKa = 11.67DD388 pKa = 3.93LAAANYY394 pKa = 8.25YY395 pKa = 10.49NSTVSVLMNLSPVCPLGYY413 pKa = 8.8STIQSLSSKK422 pKa = 10.3NINEE426 pKa = 4.07NEE428 pKa = 4.04SLTFKK433 pKa = 10.84VSVVKK438 pKa = 10.74GCTQIPVLSALNLPPNSSFFDD459 pKa = 3.49SGNGSGVFTFTPSYY473 pKa = 8.19TQSGTYY479 pKa = 9.88YY480 pKa = 9.56VTFTACDD487 pKa = 3.38TVACDD492 pKa = 3.85SEE494 pKa = 5.01TVQITVNCGTTKK506 pKa = 10.65RR507 pKa = 11.84GDD509 pKa = 3.61PNGNGQITLPDD520 pKa = 3.22VVYY523 pKa = 10.59LVNYY527 pKa = 7.69VFKK530 pKa = 10.75GGPAPVPEE538 pKa = 4.96KK539 pKa = 10.92CDD541 pKa = 3.14GDD543 pKa = 4.18ANNDD547 pKa = 3.62NQVGLIDD554 pKa = 4.93IIYY557 pKa = 8.87LANYY561 pKa = 8.62IFKK564 pKa = 10.3DD565 pKa = 3.73SPPPQPATCCLL576 pKa = 3.45
MM1 pKa = 7.42GRR3 pKa = 11.84LDD5 pKa = 3.51LSLRR9 pKa = 11.84HH10 pKa = 6.16PPSLTSGIPQGDD22 pKa = 3.53NVNLLKK28 pKa = 10.82RR29 pKa = 11.84GVKK32 pKa = 9.64KK33 pKa = 10.61LKK35 pKa = 9.87KK36 pKa = 9.33SAMGLVVILIFLLASFQIAFSQNPDD61 pKa = 3.91SIPFAPAVDD70 pKa = 3.7YY71 pKa = 11.21AVGNNPDD78 pKa = 4.92DD79 pKa = 4.39NPDD82 pKa = 4.17CIFVSDD88 pKa = 4.82LDD90 pKa = 4.04GDD92 pKa = 4.11GDD94 pKa = 3.76IDD96 pKa = 4.72LAVAMSWNDD105 pKa = 3.63SISILMNNGDD115 pKa = 4.07GTFATQISYY124 pKa = 8.77PAGSYY129 pKa = 9.09PRR131 pKa = 11.84EE132 pKa = 4.05VFGSDD137 pKa = 3.25FDD139 pKa = 5.24SDD141 pKa = 3.7GDD143 pKa = 3.65TDD145 pKa = 4.2LAIANWYY152 pKa = 8.91GNSVSILKK160 pKa = 10.62NNGDD164 pKa = 3.49GTFYY168 pKa = 11.5AKK170 pKa = 10.16VDD172 pKa = 3.66YY173 pKa = 10.46DD174 pKa = 3.72AGVAPVSVFASDD186 pKa = 4.92LDD188 pKa = 3.81FDD190 pKa = 4.8GNQDD194 pKa = 3.61FVVSKK199 pKa = 10.23QSEE202 pKa = 4.24VGVFMNKK209 pKa = 9.08GDD211 pKa = 3.57ATFEE215 pKa = 4.02PRR217 pKa = 11.84VEE219 pKa = 4.19YY220 pKa = 9.36PAGGVSVYY228 pKa = 10.53ISDD231 pKa = 4.67LDD233 pKa = 3.94GDD235 pKa = 4.33GDD237 pKa = 3.99SDD239 pKa = 4.7LAVSSTGSDD248 pKa = 3.34EE249 pKa = 5.01ISILRR254 pKa = 11.84NNGDD258 pKa = 3.22GTFFPKK264 pKa = 10.34VNYY267 pKa = 8.42LTGHH271 pKa = 5.72VPTSIFVCDD280 pKa = 3.64IDD282 pKa = 5.74GDD284 pKa = 3.99TDD286 pKa = 3.28QDD288 pKa = 3.63IIVPNFSASTISVLKK303 pKa = 10.91NNGDD307 pKa = 3.46GTFATKK313 pKa = 10.27VDD315 pKa = 3.87YY316 pKa = 8.66PTEE319 pKa = 3.97YY320 pKa = 10.67SAVKK324 pKa = 10.22IIAIDD329 pKa = 3.8LDD331 pKa = 4.02LDD333 pKa = 3.89GDD335 pKa = 3.61QDD337 pKa = 4.23IIVSHH342 pKa = 6.54EE343 pKa = 4.14NNPIISIFKK352 pKa = 10.39NNGDD356 pKa = 3.42GTFPSRR362 pKa = 11.84LNYY365 pKa = 10.23SVGAGGVTHH374 pKa = 6.71WVVAADD380 pKa = 4.02LDD382 pKa = 4.35GDD384 pKa = 4.01GDD386 pKa = 4.21YY387 pKa = 11.67DD388 pKa = 3.93LAAANYY394 pKa = 8.25YY395 pKa = 10.49NSTVSVLMNLSPVCPLGYY413 pKa = 8.8STIQSLSSKK422 pKa = 10.3NINEE426 pKa = 4.07NEE428 pKa = 4.04SLTFKK433 pKa = 10.84VSVVKK438 pKa = 10.74GCTQIPVLSALNLPPNSSFFDD459 pKa = 3.49SGNGSGVFTFTPSYY473 pKa = 8.19TQSGTYY479 pKa = 9.88YY480 pKa = 9.56VTFTACDD487 pKa = 3.38TVACDD492 pKa = 3.85SEE494 pKa = 5.01TVQITVNCGTTKK506 pKa = 10.65RR507 pKa = 11.84GDD509 pKa = 3.61PNGNGQITLPDD520 pKa = 3.22VVYY523 pKa = 10.59LVNYY527 pKa = 7.69VFKK530 pKa = 10.75GGPAPVPEE538 pKa = 4.96KK539 pKa = 10.92CDD541 pKa = 3.14GDD543 pKa = 4.18ANNDD547 pKa = 3.62NQVGLIDD554 pKa = 4.93IIYY557 pKa = 8.87LANYY561 pKa = 8.62IFKK564 pKa = 10.3DD565 pKa = 3.73SPPPQPATCCLL576 pKa = 3.45
Molecular weight: 61.27 kDa
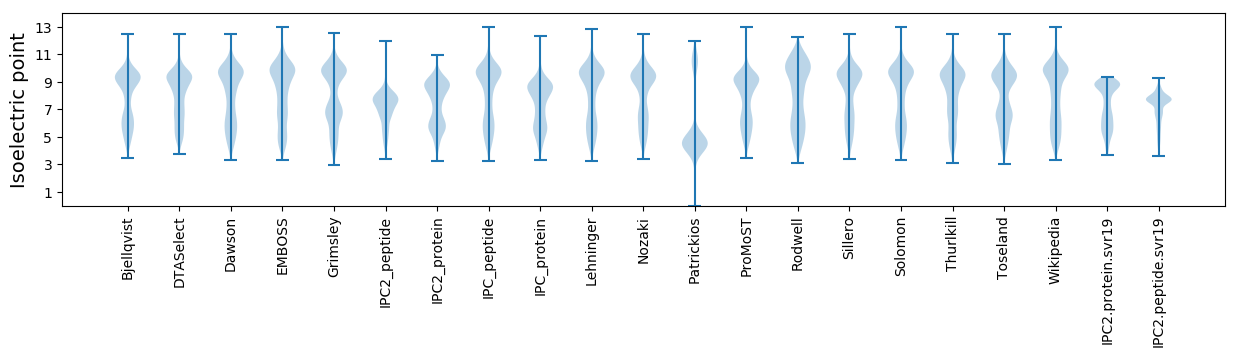
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T0LED2|T0LED2_9BACT Purine nucleoside phosphorylase OS=candidate division Zixibacteria bacterium RBG-1 OX=1379698 GN=RBG1_1C00001G0102 PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 9.98QPHH8 pKa = 4.95NRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84IKK14 pKa = 10.66DD15 pKa = 2.99HH16 pKa = 6.24GFRR19 pKa = 11.84KK20 pKa = 9.8RR21 pKa = 11.84MKK23 pKa = 8.65TKK25 pKa = 10.23GGRR28 pKa = 11.84KK29 pKa = 7.58VLRR32 pKa = 11.84KK33 pKa = 9.02RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.15GRR39 pKa = 11.84HH40 pKa = 5.44RR41 pKa = 11.84LTVTVPKK48 pKa = 10.72AA49 pKa = 3.08
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 9.98QPHH8 pKa = 4.95NRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84IKK14 pKa = 10.66DD15 pKa = 2.99HH16 pKa = 6.24GFRR19 pKa = 11.84KK20 pKa = 9.8RR21 pKa = 11.84MKK23 pKa = 8.65TKK25 pKa = 10.23GGRR28 pKa = 11.84KK29 pKa = 7.58VLRR32 pKa = 11.84KK33 pKa = 9.02RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.15GRR39 pKa = 11.84HH40 pKa = 5.44RR41 pKa = 11.84LTVTVPKK48 pKa = 10.72AA49 pKa = 3.08
Molecular weight: 6.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
641823 |
29 |
5949 |
336.7 |
37.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.815 ± 0.05 | 0.954 ± 0.028 |
4.93 ± 0.048 | 6.452 ± 0.078 |
4.947 ± 0.051 | 7.099 ± 0.063 |
1.55 ± 0.024 | 7.301 ± 0.061 |
8.082 ± 0.099 | 10.486 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.897 ± 0.032 | 4.595 ± 0.058 |
4.211 ± 0.037 | 3.559 ± 0.038 |
3.992 ± 0.038 | 6.435 ± 0.056 |
5.193 ± 0.073 | 6.861 ± 0.051 |
1.17 ± 0.021 | 3.473 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
