
Murine pneumonia virus (strain 15) (MPV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Pneumoviridae; Orthopneumovirus; Murine orthopneumovirus
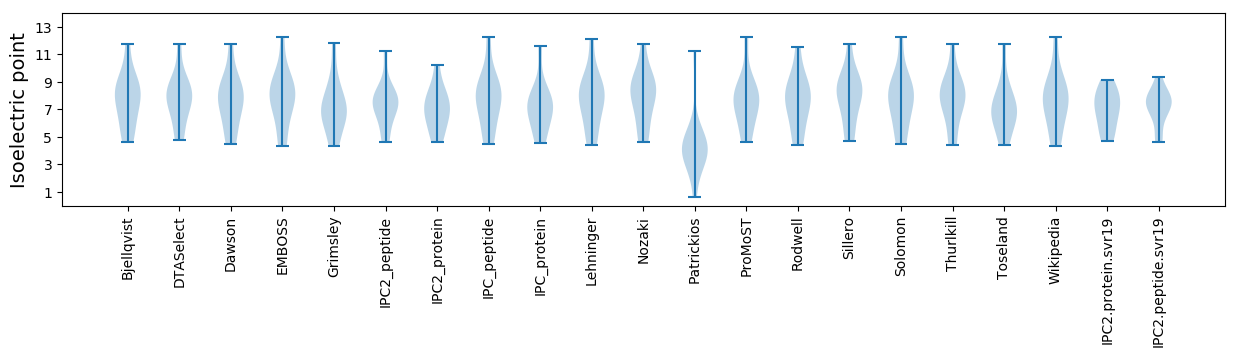
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
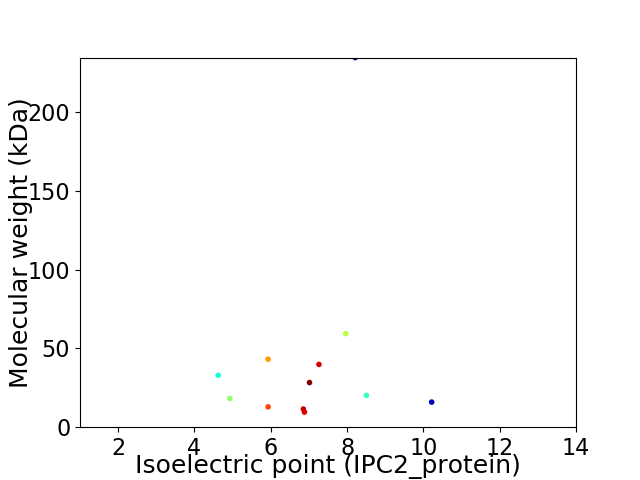
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q5MKM7|PHOSP_MPV15 Phosphoprotein OS=Murine pneumonia virus (strain 15) OX=296738 GN=P PE=3 SV=1
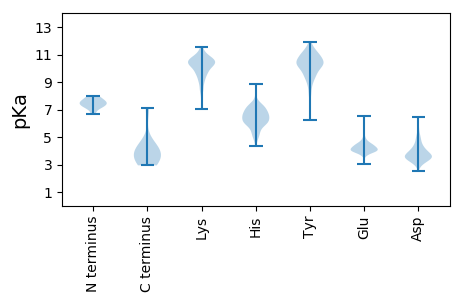
MM1 pKa = 7.59EE2 pKa = 4.81KK3 pKa = 10.16FAPEE7 pKa = 4.1FVGEE11 pKa = 4.24DD12 pKa = 3.32ANKK15 pKa = 10.21KK16 pKa = 10.09AEE18 pKa = 4.2EE19 pKa = 4.03FLKK22 pKa = 10.78HH23 pKa = 6.57RR24 pKa = 11.84SFPSEE29 pKa = 4.14KK30 pKa = 9.72PLAGIPNTATHH41 pKa = 4.6VTKK44 pKa = 11.22YY45 pKa = 10.91NMPPILRR52 pKa = 11.84SSFKK56 pKa = 10.82LPSPRR61 pKa = 11.84VAANLTEE68 pKa = 4.54PSAPPTTPPPTPPQNKK84 pKa = 9.02EE85 pKa = 4.0EE86 pKa = 4.08QPKK89 pKa = 10.61EE90 pKa = 3.97SDD92 pKa = 3.06VDD94 pKa = 3.37IEE96 pKa = 4.4TMHH99 pKa = 5.92VCKK102 pKa = 10.97VPDD105 pKa = 3.65NPEE108 pKa = 3.91HH109 pKa = 6.81SKK111 pKa = 10.94KK112 pKa = 10.2PCCSDD117 pKa = 3.33DD118 pKa = 3.54TDD120 pKa = 3.73TKK122 pKa = 10.05KK123 pKa = 8.88TRR125 pKa = 11.84KK126 pKa = 9.57PMVTFVEE133 pKa = 4.45PEE135 pKa = 3.94EE136 pKa = 4.52KK137 pKa = 10.32FVGLGASLYY146 pKa = 10.21RR147 pKa = 11.84EE148 pKa = 4.25TMQTFAADD156 pKa = 4.28GYY158 pKa = 10.96DD159 pKa = 3.57EE160 pKa = 4.49EE161 pKa = 5.77SNLSFEE167 pKa = 4.68EE168 pKa = 4.5TNQEE172 pKa = 4.04PGSSSVEE179 pKa = 3.72QRR181 pKa = 11.84LDD183 pKa = 3.74RR184 pKa = 11.84IEE186 pKa = 4.7EE187 pKa = 4.01KK188 pKa = 10.6LSYY191 pKa = 10.39IIGLLNTIMVATAGPTTARR210 pKa = 11.84DD211 pKa = 3.91EE212 pKa = 4.32IRR214 pKa = 11.84DD215 pKa = 3.73ALIGTRR221 pKa = 11.84EE222 pKa = 4.02EE223 pKa = 4.52LIEE226 pKa = 4.22MIKK229 pKa = 10.57SDD231 pKa = 3.56ILTVNDD237 pKa = 4.83RR238 pKa = 11.84IVAMEE243 pKa = 3.95KK244 pKa = 10.61LRR246 pKa = 11.84DD247 pKa = 3.97EE248 pKa = 4.61EE249 pKa = 4.44CSRR252 pKa = 11.84ADD254 pKa = 3.4TDD256 pKa = 5.52DD257 pKa = 4.83GSACYY262 pKa = 8.95LTDD265 pKa = 3.61RR266 pKa = 11.84ARR268 pKa = 11.84ILDD271 pKa = 4.28KK272 pKa = 10.67IVSSNAEE279 pKa = 3.88EE280 pKa = 4.12AKK282 pKa = 10.64EE283 pKa = 4.12DD284 pKa = 4.47LDD286 pKa = 3.84VDD288 pKa = 4.74DD289 pKa = 5.61IMGINFF295 pKa = 3.75
MM1 pKa = 7.59EE2 pKa = 4.81KK3 pKa = 10.16FAPEE7 pKa = 4.1FVGEE11 pKa = 4.24DD12 pKa = 3.32ANKK15 pKa = 10.21KK16 pKa = 10.09AEE18 pKa = 4.2EE19 pKa = 4.03FLKK22 pKa = 10.78HH23 pKa = 6.57RR24 pKa = 11.84SFPSEE29 pKa = 4.14KK30 pKa = 9.72PLAGIPNTATHH41 pKa = 4.6VTKK44 pKa = 11.22YY45 pKa = 10.91NMPPILRR52 pKa = 11.84SSFKK56 pKa = 10.82LPSPRR61 pKa = 11.84VAANLTEE68 pKa = 4.54PSAPPTTPPPTPPQNKK84 pKa = 9.02EE85 pKa = 4.0EE86 pKa = 4.08QPKK89 pKa = 10.61EE90 pKa = 3.97SDD92 pKa = 3.06VDD94 pKa = 3.37IEE96 pKa = 4.4TMHH99 pKa = 5.92VCKK102 pKa = 10.97VPDD105 pKa = 3.65NPEE108 pKa = 3.91HH109 pKa = 6.81SKK111 pKa = 10.94KK112 pKa = 10.2PCCSDD117 pKa = 3.33DD118 pKa = 3.54TDD120 pKa = 3.73TKK122 pKa = 10.05KK123 pKa = 8.88TRR125 pKa = 11.84KK126 pKa = 9.57PMVTFVEE133 pKa = 4.45PEE135 pKa = 3.94EE136 pKa = 4.52KK137 pKa = 10.32FVGLGASLYY146 pKa = 10.21RR147 pKa = 11.84EE148 pKa = 4.25TMQTFAADD156 pKa = 4.28GYY158 pKa = 10.96DD159 pKa = 3.57EE160 pKa = 4.49EE161 pKa = 5.77SNLSFEE167 pKa = 4.68EE168 pKa = 4.5TNQEE172 pKa = 4.04PGSSSVEE179 pKa = 3.72QRR181 pKa = 11.84LDD183 pKa = 3.74RR184 pKa = 11.84IEE186 pKa = 4.7EE187 pKa = 4.01KK188 pKa = 10.6LSYY191 pKa = 10.39IIGLLNTIMVATAGPTTARR210 pKa = 11.84DD211 pKa = 3.91EE212 pKa = 4.32IRR214 pKa = 11.84DD215 pKa = 3.73ALIGTRR221 pKa = 11.84EE222 pKa = 4.02EE223 pKa = 4.52LIEE226 pKa = 4.22MIKK229 pKa = 10.57SDD231 pKa = 3.56ILTVNDD237 pKa = 4.83RR238 pKa = 11.84IVAMEE243 pKa = 3.95KK244 pKa = 10.61LRR246 pKa = 11.84DD247 pKa = 3.97EE248 pKa = 4.61EE249 pKa = 4.44CSRR252 pKa = 11.84ADD254 pKa = 3.4TDD256 pKa = 5.52DD257 pKa = 4.83GSACYY262 pKa = 8.95LTDD265 pKa = 3.61RR266 pKa = 11.84ARR268 pKa = 11.84ILDD271 pKa = 4.28KK272 pKa = 10.67IVSSNAEE279 pKa = 3.88EE280 pKa = 4.12AKK282 pKa = 10.64EE283 pKa = 4.12DD284 pKa = 4.47LDD286 pKa = 3.84VDD288 pKa = 4.74DD289 pKa = 5.61IMGINFF295 pKa = 3.75
Molecular weight: 32.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q5MKM7|PHOSP_MPV15 Phosphoprotein OS=Murine pneumonia virus (strain 15) OX=296738 GN=P PE=3 SV=1
MM1 pKa = 7.5SPNITCPLYY10 pKa = 10.92CVAHH14 pKa = 6.3SNSLPRR20 pKa = 11.84EE21 pKa = 3.91LLQILLNPLLPLPLHH36 pKa = 6.52HH37 pKa = 7.06PHH39 pKa = 7.28LPRR42 pKa = 11.84TRR44 pKa = 11.84KK45 pKa = 9.7SSPKK49 pKa = 10.59SLMLTLRR56 pKa = 11.84LCMSVRR62 pKa = 11.84FLTIRR67 pKa = 11.84NTARR71 pKa = 11.84SHH73 pKa = 5.98AAQMIPILRR82 pKa = 11.84KK83 pKa = 9.87LGSRR87 pKa = 11.84WSPLWNPRR95 pKa = 11.84RR96 pKa = 11.84NLSDD100 pKa = 3.3WEE102 pKa = 4.26LACTGRR108 pKa = 11.84PCRR111 pKa = 11.84PLLLMVMMKK120 pKa = 10.2KK121 pKa = 9.03ATYY124 pKa = 10.09RR125 pKa = 11.84LRR127 pKa = 11.84RR128 pKa = 11.84LTKK131 pKa = 10.42SRR133 pKa = 11.84VLHH136 pKa = 6.18LL137 pKa = 4.29
MM1 pKa = 7.5SPNITCPLYY10 pKa = 10.92CVAHH14 pKa = 6.3SNSLPRR20 pKa = 11.84EE21 pKa = 3.91LLQILLNPLLPLPLHH36 pKa = 6.52HH37 pKa = 7.06PHH39 pKa = 7.28LPRR42 pKa = 11.84TRR44 pKa = 11.84KK45 pKa = 9.7SSPKK49 pKa = 10.59SLMLTLRR56 pKa = 11.84LCMSVRR62 pKa = 11.84FLTIRR67 pKa = 11.84NTARR71 pKa = 11.84SHH73 pKa = 5.98AAQMIPILRR82 pKa = 11.84KK83 pKa = 9.87LGSRR87 pKa = 11.84WSPLWNPRR95 pKa = 11.84RR96 pKa = 11.84NLSDD100 pKa = 3.3WEE102 pKa = 4.26LACTGRR108 pKa = 11.84PCRR111 pKa = 11.84PLLLMVMMKK120 pKa = 10.2KK121 pKa = 9.03ATYY124 pKa = 10.09RR125 pKa = 11.84LRR127 pKa = 11.84RR128 pKa = 11.84LTKK131 pKa = 10.42SRR133 pKa = 11.84VLHH136 pKa = 6.18LL137 pKa = 4.29
Molecular weight: 15.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4657 |
92 |
2040 |
388.1 |
43.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.239 ± 0.933 | 2.319 ± 0.365 |
5.132 ± 0.366 | 5.068 ± 0.556 |
3.457 ± 0.395 | 4.853 ± 0.411 |
2.491 ± 0.265 | 6.764 ± 0.516 |
6.141 ± 0.358 | 10.994 ± 0.878 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.791 ± 0.196 | 5.841 ± 0.307 |
4.531 ± 0.753 | 3.028 ± 0.301 |
5.175 ± 0.413 | 8.267 ± 0.394 |
6.657 ± 0.691 | 6.335 ± 0.561 |
1.16 ± 0.243 | 3.758 ± 0.341 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
