
Beihai sipunculid worm virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
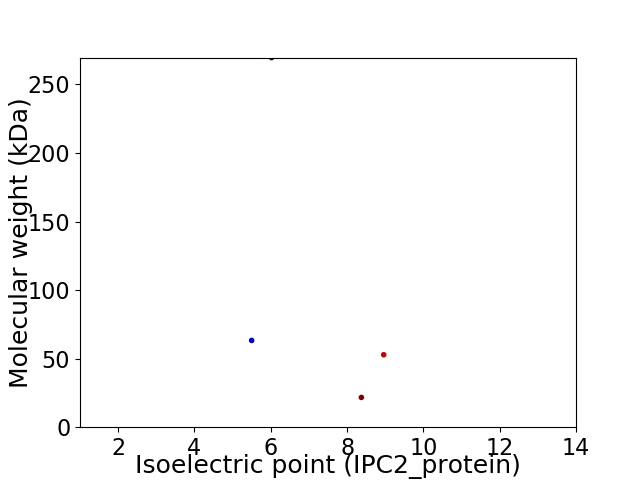
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHN2|A0A1L3KHN2_9VIRU Uncharacterized protein OS=Beihai sipunculid worm virus 5 OX=1922677 PE=4 SV=1
MM1 pKa = 7.63SMPATATGTPKK12 pKa = 10.76NYY14 pKa = 10.07DD15 pKa = 2.85VSAQGGGPAALDD27 pKa = 3.72TQQVPVEE34 pKa = 4.27ASQTAIVAHH43 pKa = 6.28SGDD46 pKa = 3.78VNVIDD51 pKa = 5.31PYY53 pKa = 10.8LKK55 pKa = 10.36KK56 pKa = 10.16HH57 pKa = 6.67FILNSIFRR65 pKa = 11.84WTTAMPAGTIIRR77 pKa = 11.84KK78 pKa = 9.2IPVHH82 pKa = 6.27PSYY85 pKa = 11.54ANPQVQHH92 pKa = 6.67LSQMYY97 pKa = 9.1NLWAGSLDD105 pKa = 3.77YY106 pKa = 10.66KK107 pKa = 10.81IEE109 pKa = 3.87IAGTGFHH116 pKa = 6.99GGRR119 pKa = 11.84LLVAKK124 pKa = 9.49VPPNLNPDD132 pKa = 3.68DD133 pKa = 4.16YY134 pKa = 10.32TPQGLTVFHH143 pKa = 6.26NVEE146 pKa = 4.07IEE148 pKa = 4.35VKK150 pKa = 10.13QSMGGTLTAIDD161 pKa = 3.65QKK163 pKa = 11.41NIVYY167 pKa = 9.89HH168 pKa = 5.71YY169 pKa = 10.73NSGSKK174 pKa = 10.29PEE176 pKa = 4.57LGSVDD181 pKa = 3.52GGTLVIIVLYY191 pKa = 9.42PLVLAAEE198 pKa = 4.52QNGSVNLGLYY208 pKa = 10.23SKK210 pKa = 10.91LGDD213 pKa = 3.77DD214 pKa = 4.42FVFSQMQPVLQTTTYY229 pKa = 11.22NPTHH233 pKa = 6.83LNNMFRR239 pKa = 11.84FDD241 pKa = 4.22HH242 pKa = 6.76RR243 pKa = 11.84EE244 pKa = 3.84CLHH247 pKa = 6.38PVVTACVFKK256 pKa = 10.9KK257 pKa = 10.65LRR259 pKa = 11.84VSKK262 pKa = 9.88NQKK265 pKa = 8.51EE266 pKa = 4.73VYY268 pKa = 9.87HH269 pKa = 6.21GLGFLAGADD278 pKa = 3.42DD279 pKa = 3.66TRR281 pKa = 11.84RR282 pKa = 11.84LFKK285 pKa = 9.88EE286 pKa = 3.69HH287 pKa = 7.14RR288 pKa = 11.84NDD290 pKa = 3.71TRR292 pKa = 11.84GCSNTDD298 pKa = 2.96GTSATPVQGFTRR310 pKa = 11.84IFEE313 pKa = 4.39SQVQFGLNIYY323 pKa = 10.04HH324 pKa = 6.69NGGNGVTDD332 pKa = 3.24QVYY335 pKa = 7.99TAQGNVSNPAKK346 pKa = 10.36GYY348 pKa = 9.63VWLQHH353 pKa = 6.56GIHH356 pKa = 7.16DD357 pKa = 4.34AVQNEE362 pKa = 3.93NAYY365 pKa = 10.04CAEE368 pKa = 4.2GTKK371 pKa = 10.18SHH373 pKa = 7.42LDD375 pKa = 3.74PIAWYY380 pKa = 9.18EE381 pKa = 4.66DD382 pKa = 4.08PLCPPPAPVSTTNGEE397 pKa = 4.29GIVYY401 pKa = 8.37FQFDD405 pKa = 3.63GDD407 pKa = 4.0TTDD410 pKa = 4.16RR411 pKa = 11.84GCLQDD416 pKa = 3.72YY417 pKa = 9.32TIMSMFRR424 pKa = 11.84AGALEE429 pKa = 3.75EE430 pKa = 4.22LALGEE435 pKa = 4.38SVVFQLVDD443 pKa = 3.07ARR445 pKa = 11.84TNNVLNYY452 pKa = 9.8VRR454 pKa = 11.84LHH456 pKa = 6.57RR457 pKa = 11.84EE458 pKa = 3.76GFMSAVMPATSTEE471 pKa = 3.6YY472 pKa = 10.72DD473 pKa = 3.33LRR475 pKa = 11.84DD476 pKa = 2.91IYY478 pKa = 10.87LRR480 pKa = 11.84YY481 pKa = 9.08MEE483 pKa = 5.36KK484 pKa = 10.78LPVSNPLPGSQLAEE498 pKa = 3.9LTSQNYY504 pKa = 9.96ALNKK508 pKa = 10.06AADD511 pKa = 3.78ALRR514 pKa = 11.84DD515 pKa = 3.39EE516 pKa = 4.54LMEE519 pKa = 4.23VKK521 pKa = 10.18QAFAGLASSVEE532 pKa = 4.44SEE534 pKa = 4.19LQTSKK539 pKa = 10.97GSKK542 pKa = 10.01LPWRR546 pKa = 11.84RR547 pKa = 11.84QQQKK551 pKa = 9.78QSQQRR556 pKa = 11.84LQGALAASLEE566 pKa = 4.32SLEE569 pKa = 4.44EE570 pKa = 4.07TSSLQAA576 pKa = 4.09
MM1 pKa = 7.63SMPATATGTPKK12 pKa = 10.76NYY14 pKa = 10.07DD15 pKa = 2.85VSAQGGGPAALDD27 pKa = 3.72TQQVPVEE34 pKa = 4.27ASQTAIVAHH43 pKa = 6.28SGDD46 pKa = 3.78VNVIDD51 pKa = 5.31PYY53 pKa = 10.8LKK55 pKa = 10.36KK56 pKa = 10.16HH57 pKa = 6.67FILNSIFRR65 pKa = 11.84WTTAMPAGTIIRR77 pKa = 11.84KK78 pKa = 9.2IPVHH82 pKa = 6.27PSYY85 pKa = 11.54ANPQVQHH92 pKa = 6.67LSQMYY97 pKa = 9.1NLWAGSLDD105 pKa = 3.77YY106 pKa = 10.66KK107 pKa = 10.81IEE109 pKa = 3.87IAGTGFHH116 pKa = 6.99GGRR119 pKa = 11.84LLVAKK124 pKa = 9.49VPPNLNPDD132 pKa = 3.68DD133 pKa = 4.16YY134 pKa = 10.32TPQGLTVFHH143 pKa = 6.26NVEE146 pKa = 4.07IEE148 pKa = 4.35VKK150 pKa = 10.13QSMGGTLTAIDD161 pKa = 3.65QKK163 pKa = 11.41NIVYY167 pKa = 9.89HH168 pKa = 5.71YY169 pKa = 10.73NSGSKK174 pKa = 10.29PEE176 pKa = 4.57LGSVDD181 pKa = 3.52GGTLVIIVLYY191 pKa = 9.42PLVLAAEE198 pKa = 4.52QNGSVNLGLYY208 pKa = 10.23SKK210 pKa = 10.91LGDD213 pKa = 3.77DD214 pKa = 4.42FVFSQMQPVLQTTTYY229 pKa = 11.22NPTHH233 pKa = 6.83LNNMFRR239 pKa = 11.84FDD241 pKa = 4.22HH242 pKa = 6.76RR243 pKa = 11.84EE244 pKa = 3.84CLHH247 pKa = 6.38PVVTACVFKK256 pKa = 10.9KK257 pKa = 10.65LRR259 pKa = 11.84VSKK262 pKa = 9.88NQKK265 pKa = 8.51EE266 pKa = 4.73VYY268 pKa = 9.87HH269 pKa = 6.21GLGFLAGADD278 pKa = 3.42DD279 pKa = 3.66TRR281 pKa = 11.84RR282 pKa = 11.84LFKK285 pKa = 9.88EE286 pKa = 3.69HH287 pKa = 7.14RR288 pKa = 11.84NDD290 pKa = 3.71TRR292 pKa = 11.84GCSNTDD298 pKa = 2.96GTSATPVQGFTRR310 pKa = 11.84IFEE313 pKa = 4.39SQVQFGLNIYY323 pKa = 10.04HH324 pKa = 6.69NGGNGVTDD332 pKa = 3.24QVYY335 pKa = 7.99TAQGNVSNPAKK346 pKa = 10.36GYY348 pKa = 9.63VWLQHH353 pKa = 6.56GIHH356 pKa = 7.16DD357 pKa = 4.34AVQNEE362 pKa = 3.93NAYY365 pKa = 10.04CAEE368 pKa = 4.2GTKK371 pKa = 10.18SHH373 pKa = 7.42LDD375 pKa = 3.74PIAWYY380 pKa = 9.18EE381 pKa = 4.66DD382 pKa = 4.08PLCPPPAPVSTTNGEE397 pKa = 4.29GIVYY401 pKa = 8.37FQFDD405 pKa = 3.63GDD407 pKa = 4.0TTDD410 pKa = 4.16RR411 pKa = 11.84GCLQDD416 pKa = 3.72YY417 pKa = 9.32TIMSMFRR424 pKa = 11.84AGALEE429 pKa = 3.75EE430 pKa = 4.22LALGEE435 pKa = 4.38SVVFQLVDD443 pKa = 3.07ARR445 pKa = 11.84TNNVLNYY452 pKa = 9.8VRR454 pKa = 11.84LHH456 pKa = 6.57RR457 pKa = 11.84EE458 pKa = 3.76GFMSAVMPATSTEE471 pKa = 3.6YY472 pKa = 10.72DD473 pKa = 3.33LRR475 pKa = 11.84DD476 pKa = 2.91IYY478 pKa = 10.87LRR480 pKa = 11.84YY481 pKa = 9.08MEE483 pKa = 5.36KK484 pKa = 10.78LPVSNPLPGSQLAEE498 pKa = 3.9LTSQNYY504 pKa = 9.96ALNKK508 pKa = 10.06AADD511 pKa = 3.78ALRR514 pKa = 11.84DD515 pKa = 3.39EE516 pKa = 4.54LMEE519 pKa = 4.23VKK521 pKa = 10.18QAFAGLASSVEE532 pKa = 4.44SEE534 pKa = 4.19LQTSKK539 pKa = 10.97GSKK542 pKa = 10.01LPWRR546 pKa = 11.84RR547 pKa = 11.84QQQKK551 pKa = 9.78QSQQRR556 pKa = 11.84LQGALAASLEE566 pKa = 4.32SLEE569 pKa = 4.44EE570 pKa = 4.07TSSLQAA576 pKa = 4.09
Molecular weight: 63.29 kDa
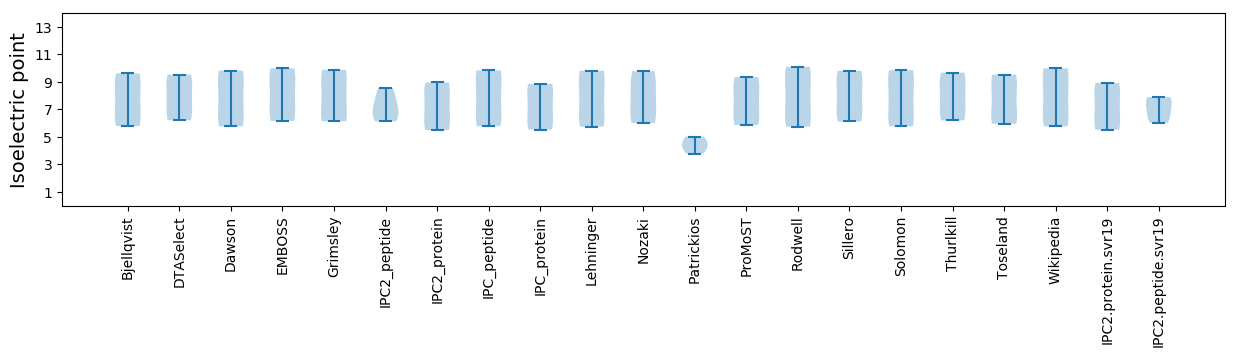
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHK3|A0A1L3KHK3_9VIRU Calici_coat domain-containing protein OS=Beihai sipunculid worm virus 5 OX=1922677 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 4.78SANLSISPVYY12 pKa = 10.26QGPLDD17 pKa = 4.46LNSEE21 pKa = 4.39IYY23 pKa = 10.75VGEE26 pKa = 4.1CDD28 pKa = 4.15KK29 pKa = 11.58FGDD32 pKa = 3.39KK33 pKa = 10.19RR34 pKa = 11.84KK35 pKa = 10.1RR36 pKa = 11.84RR37 pKa = 11.84VKK39 pKa = 10.17IKK41 pKa = 10.4NVRR44 pKa = 11.84CKK46 pKa = 10.66QIVHH50 pKa = 5.56TVIFGNTEE58 pKa = 3.58IPVTLACEE66 pKa = 4.17EE67 pKa = 3.96NRR69 pKa = 11.84QAVHH73 pKa = 7.19ANTCPHH79 pKa = 6.3GNEE82 pKa = 3.86TVPRR86 pKa = 11.84LKK88 pKa = 10.51CIAAARR94 pKa = 11.84VFEE97 pKa = 4.32SSRR100 pKa = 11.84LEE102 pKa = 3.91PPPAISRR109 pKa = 11.84FQKK112 pKa = 9.28EE113 pKa = 3.54NCYY116 pKa = 9.73FASQVFRR123 pKa = 11.84LFCFILQCIPSLVTMLPPEE142 pKa = 4.64SGPSVKK148 pKa = 10.28VLEE151 pKa = 4.44SEE153 pKa = 4.23KK154 pKa = 10.78VPLFTRR160 pKa = 11.84YY161 pKa = 9.11HH162 pKa = 6.34LKK164 pKa = 10.73SMPDD168 pKa = 3.55RR169 pKa = 11.84TPMDD173 pKa = 4.16EE174 pKa = 6.1DD175 pKa = 3.12ISTYY179 pKa = 9.15IWCVQPIHH187 pKa = 6.39IWVFEE192 pKa = 4.02LVVAHH197 pKa = 6.41INWLRR202 pKa = 11.84FCRR205 pKa = 11.84FPSVYY210 pKa = 9.33TYY212 pKa = 11.42LSGTCLCSQAPFFICSDD229 pKa = 3.7EE230 pKa = 4.18KK231 pKa = 11.02CRR233 pKa = 11.84RR234 pKa = 11.84NPKK237 pKa = 8.54TDD239 pKa = 3.33FRR241 pKa = 11.84FWPRR245 pKa = 11.84FVPQEE250 pKa = 3.98LSTDD254 pKa = 3.44RR255 pKa = 11.84LDD257 pKa = 4.3EE258 pKa = 4.35LKK260 pKa = 10.47QYY262 pKa = 10.66KK263 pKa = 6.5WTRR266 pKa = 11.84SLRR269 pKa = 11.84AGVWRR274 pKa = 11.84MLSGQRR280 pKa = 11.84RR281 pKa = 11.84FIKK284 pKa = 10.27PMIPSEE290 pKa = 4.15IEE292 pKa = 3.58SSYY295 pKa = 10.7MKK297 pKa = 10.41VIQFIYY303 pKa = 8.32WQSFLLCSYY312 pKa = 10.05GRR314 pKa = 11.84RR315 pKa = 11.84AHH317 pKa = 6.18PHH319 pKa = 4.28NHH321 pKa = 6.13PPVKK325 pKa = 10.48NVMLWRR331 pKa = 11.84TKK333 pKa = 9.4TKK335 pKa = 10.73LVGSFINYY343 pKa = 5.87MWTNFGSLPRR353 pKa = 11.84KK354 pKa = 9.09EE355 pKa = 4.95DD356 pKa = 3.31LVPFYY361 pKa = 11.2RR362 pKa = 11.84MTKK365 pKa = 7.96MMRR368 pKa = 11.84TKK370 pKa = 10.43FYY372 pKa = 11.45AEE374 pKa = 3.85VLAVKK379 pKa = 9.75QKK381 pKa = 10.42KK382 pKa = 9.98GKK384 pKa = 8.44ATEE387 pKa = 4.13SDD389 pKa = 2.72RR390 pKa = 11.84HH391 pKa = 5.92LIRR394 pKa = 11.84RR395 pKa = 11.84SLHH398 pKa = 4.96QIWYY402 pKa = 9.29KK403 pKa = 10.52VLSPSTPNKK412 pKa = 10.04LYY414 pKa = 10.64NDD416 pKa = 4.3RR417 pKa = 11.84ASHH420 pKa = 6.3LAAVLIEE427 pKa = 4.29YY428 pKa = 10.34INRR431 pKa = 11.84LPFLKK436 pKa = 10.71SLGQEE441 pKa = 3.55KK442 pKa = 10.54CFNIVLNFSS451 pKa = 3.54
MM1 pKa = 8.1DD2 pKa = 4.78SANLSISPVYY12 pKa = 10.26QGPLDD17 pKa = 4.46LNSEE21 pKa = 4.39IYY23 pKa = 10.75VGEE26 pKa = 4.1CDD28 pKa = 4.15KK29 pKa = 11.58FGDD32 pKa = 3.39KK33 pKa = 10.19RR34 pKa = 11.84KK35 pKa = 10.1RR36 pKa = 11.84RR37 pKa = 11.84VKK39 pKa = 10.17IKK41 pKa = 10.4NVRR44 pKa = 11.84CKK46 pKa = 10.66QIVHH50 pKa = 5.56TVIFGNTEE58 pKa = 3.58IPVTLACEE66 pKa = 4.17EE67 pKa = 3.96NRR69 pKa = 11.84QAVHH73 pKa = 7.19ANTCPHH79 pKa = 6.3GNEE82 pKa = 3.86TVPRR86 pKa = 11.84LKK88 pKa = 10.51CIAAARR94 pKa = 11.84VFEE97 pKa = 4.32SSRR100 pKa = 11.84LEE102 pKa = 3.91PPPAISRR109 pKa = 11.84FQKK112 pKa = 9.28EE113 pKa = 3.54NCYY116 pKa = 9.73FASQVFRR123 pKa = 11.84LFCFILQCIPSLVTMLPPEE142 pKa = 4.64SGPSVKK148 pKa = 10.28VLEE151 pKa = 4.44SEE153 pKa = 4.23KK154 pKa = 10.78VPLFTRR160 pKa = 11.84YY161 pKa = 9.11HH162 pKa = 6.34LKK164 pKa = 10.73SMPDD168 pKa = 3.55RR169 pKa = 11.84TPMDD173 pKa = 4.16EE174 pKa = 6.1DD175 pKa = 3.12ISTYY179 pKa = 9.15IWCVQPIHH187 pKa = 6.39IWVFEE192 pKa = 4.02LVVAHH197 pKa = 6.41INWLRR202 pKa = 11.84FCRR205 pKa = 11.84FPSVYY210 pKa = 9.33TYY212 pKa = 11.42LSGTCLCSQAPFFICSDD229 pKa = 3.7EE230 pKa = 4.18KK231 pKa = 11.02CRR233 pKa = 11.84RR234 pKa = 11.84NPKK237 pKa = 8.54TDD239 pKa = 3.33FRR241 pKa = 11.84FWPRR245 pKa = 11.84FVPQEE250 pKa = 3.98LSTDD254 pKa = 3.44RR255 pKa = 11.84LDD257 pKa = 4.3EE258 pKa = 4.35LKK260 pKa = 10.47QYY262 pKa = 10.66KK263 pKa = 6.5WTRR266 pKa = 11.84SLRR269 pKa = 11.84AGVWRR274 pKa = 11.84MLSGQRR280 pKa = 11.84RR281 pKa = 11.84FIKK284 pKa = 10.27PMIPSEE290 pKa = 4.15IEE292 pKa = 3.58SSYY295 pKa = 10.7MKK297 pKa = 10.41VIQFIYY303 pKa = 8.32WQSFLLCSYY312 pKa = 10.05GRR314 pKa = 11.84RR315 pKa = 11.84AHH317 pKa = 6.18PHH319 pKa = 4.28NHH321 pKa = 6.13PPVKK325 pKa = 10.48NVMLWRR331 pKa = 11.84TKK333 pKa = 9.4TKK335 pKa = 10.73LVGSFINYY343 pKa = 5.87MWTNFGSLPRR353 pKa = 11.84KK354 pKa = 9.09EE355 pKa = 4.95DD356 pKa = 3.31LVPFYY361 pKa = 11.2RR362 pKa = 11.84MTKK365 pKa = 7.96MMRR368 pKa = 11.84TKK370 pKa = 10.43FYY372 pKa = 11.45AEE374 pKa = 3.85VLAVKK379 pKa = 9.75QKK381 pKa = 10.42KK382 pKa = 9.98GKK384 pKa = 8.44ATEE387 pKa = 4.13SDD389 pKa = 2.72RR390 pKa = 11.84HH391 pKa = 5.92LIRR394 pKa = 11.84RR395 pKa = 11.84SLHH398 pKa = 4.96QIWYY402 pKa = 9.29KK403 pKa = 10.52VLSPSTPNKK412 pKa = 10.04LYY414 pKa = 10.64NDD416 pKa = 4.3RR417 pKa = 11.84ASHH420 pKa = 6.3LAAVLIEE427 pKa = 4.29YY428 pKa = 10.34INRR431 pKa = 11.84LPFLKK436 pKa = 10.71SLGQEE441 pKa = 3.55KK442 pKa = 10.54CFNIVLNFSS451 pKa = 3.54
Molecular weight: 52.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3597 |
202 |
2368 |
899.3 |
101.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.617 ± 0.702 | 1.974 ± 0.397 |
5.366 ± 0.648 | 6.839 ± 1.01 |
4.198 ± 0.392 | 6.144 ± 0.821 |
2.585 ± 0.292 | 4.754 ± 0.355 |
7.145 ± 0.952 | 8.201 ± 0.542 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.613 ± 0.198 | 4.448 ± 0.55 |
4.643 ± 0.575 | 4.865 ± 0.591 |
5.032 ± 0.601 | 6.533 ± 0.416 |
6.339 ± 0.494 | 6.533 ± 0.385 |
1.501 ± 0.207 | 3.67 ± 0.221 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
