
Plantago lanceolata latent virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Capulavirus
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
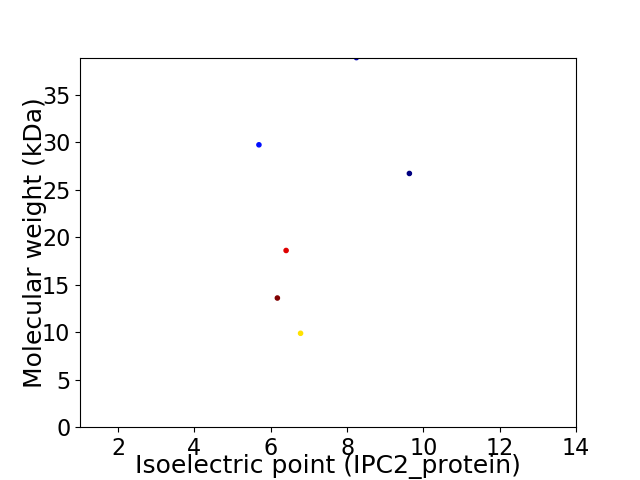
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A166V5R3|A0A166V5R3_9GEMI Replication-associated protein OS=Plantago lanceolata latent virus OX=1830242 PE=3 SV=1
MM1 pKa = 7.62PRR3 pKa = 11.84QPLTYY8 pKa = 10.0RR9 pKa = 11.84FQGKK13 pKa = 8.62SAFLTYY19 pKa = 9.2PKK21 pKa = 10.34CDD23 pKa = 3.57LTPTSVLDD31 pKa = 3.44YY32 pKa = 10.88LYY34 pKa = 11.3NLLKK38 pKa = 10.91NYY40 pKa = 10.16DD41 pKa = 2.97IFYY44 pKa = 10.99ARR46 pKa = 11.84VCQEE50 pKa = 3.42NHH52 pKa = 5.66QDD54 pKa = 3.62GTKK57 pKa = 10.14HH58 pKa = 5.53LHH60 pKa = 6.69CLVQLAKK67 pKa = 10.69RR68 pKa = 11.84LTTRR72 pKa = 11.84DD73 pKa = 3.39PKK75 pKa = 11.36YY76 pKa = 10.98FDD78 pKa = 2.88ITTANTITSGDD89 pKa = 3.95AQPHH93 pKa = 5.96HH94 pKa = 7.34PNIQVPRR101 pKa = 11.84SDD103 pKa = 3.46AHH105 pKa = 5.36VADD108 pKa = 5.39YY109 pKa = 10.38IAKK112 pKa = 10.19DD113 pKa = 3.47GNFVEE118 pKa = 4.93RR119 pKa = 11.84GVLKK123 pKa = 9.79ATRR126 pKa = 11.84RR127 pKa = 11.84SPKK130 pKa = 9.83KK131 pKa = 10.49SRR133 pKa = 11.84DD134 pKa = 4.09TIWTAILQEE143 pKa = 4.52STSKK147 pKa = 10.73PDD149 pKa = 3.22FLARR153 pKa = 11.84VQSEE157 pKa = 4.23QPYY160 pKa = 8.52TWATQLRR167 pKa = 11.84NLEE170 pKa = 4.05YY171 pKa = 10.28AANSRR176 pKa = 11.84WPEE179 pKa = 3.72PPVLYY184 pKa = 9.99EE185 pKa = 3.41PRR187 pKa = 11.84YY188 pKa = 9.56RR189 pKa = 11.84AFPNLPEE196 pKa = 5.47PIKK199 pKa = 10.65QWAEE203 pKa = 3.85TNLYY207 pKa = 8.41TVSFNSMQLIDD218 pKa = 4.76PDD220 pKa = 4.16IEE222 pKa = 4.46HH223 pKa = 7.61ADD225 pKa = 4.19LDD227 pKa = 4.22WASNTAQEE235 pKa = 4.25EE236 pKa = 4.47LEE238 pKa = 4.18EE239 pKa = 3.93QLLLNSQQPITVSDD253 pKa = 4.43AGPQTT258 pKa = 3.5
MM1 pKa = 7.62PRR3 pKa = 11.84QPLTYY8 pKa = 10.0RR9 pKa = 11.84FQGKK13 pKa = 8.62SAFLTYY19 pKa = 9.2PKK21 pKa = 10.34CDD23 pKa = 3.57LTPTSVLDD31 pKa = 3.44YY32 pKa = 10.88LYY34 pKa = 11.3NLLKK38 pKa = 10.91NYY40 pKa = 10.16DD41 pKa = 2.97IFYY44 pKa = 10.99ARR46 pKa = 11.84VCQEE50 pKa = 3.42NHH52 pKa = 5.66QDD54 pKa = 3.62GTKK57 pKa = 10.14HH58 pKa = 5.53LHH60 pKa = 6.69CLVQLAKK67 pKa = 10.69RR68 pKa = 11.84LTTRR72 pKa = 11.84DD73 pKa = 3.39PKK75 pKa = 11.36YY76 pKa = 10.98FDD78 pKa = 2.88ITTANTITSGDD89 pKa = 3.95AQPHH93 pKa = 5.96HH94 pKa = 7.34PNIQVPRR101 pKa = 11.84SDD103 pKa = 3.46AHH105 pKa = 5.36VADD108 pKa = 5.39YY109 pKa = 10.38IAKK112 pKa = 10.19DD113 pKa = 3.47GNFVEE118 pKa = 4.93RR119 pKa = 11.84GVLKK123 pKa = 9.79ATRR126 pKa = 11.84RR127 pKa = 11.84SPKK130 pKa = 9.83KK131 pKa = 10.49SRR133 pKa = 11.84DD134 pKa = 4.09TIWTAILQEE143 pKa = 4.52STSKK147 pKa = 10.73PDD149 pKa = 3.22FLARR153 pKa = 11.84VQSEE157 pKa = 4.23QPYY160 pKa = 8.52TWATQLRR167 pKa = 11.84NLEE170 pKa = 4.05YY171 pKa = 10.28AANSRR176 pKa = 11.84WPEE179 pKa = 3.72PPVLYY184 pKa = 9.99EE185 pKa = 3.41PRR187 pKa = 11.84YY188 pKa = 9.56RR189 pKa = 11.84AFPNLPEE196 pKa = 5.47PIKK199 pKa = 10.65QWAEE203 pKa = 3.85TNLYY207 pKa = 8.41TVSFNSMQLIDD218 pKa = 4.76PDD220 pKa = 4.16IEE222 pKa = 4.46HH223 pKa = 7.61ADD225 pKa = 4.19LDD227 pKa = 4.22WASNTAQEE235 pKa = 4.25EE236 pKa = 4.47LEE238 pKa = 4.18EE239 pKa = 3.93QLLLNSQQPITVSDD253 pKa = 4.43AGPQTT258 pKa = 3.5
Molecular weight: 29.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166V5P1|A0A166V5P1_9GEMI C3 OS=Plantago lanceolata latent virus OX=1830242 PE=4 SV=1
MM1 pKa = 7.38VKK3 pKa = 10.13RR4 pKa = 11.84KK5 pKa = 9.88RR6 pKa = 11.84STGVTTTYY14 pKa = 11.07AKK16 pKa = 10.26RR17 pKa = 11.84SRR19 pKa = 11.84VGSGYY24 pKa = 9.23RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84WSRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84IVRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.55GVYY41 pKa = 9.24ARR43 pKa = 11.84HH44 pKa = 6.21RR45 pKa = 11.84SQSVLHH51 pKa = 6.16SLSNATATKK60 pKa = 10.46GKK62 pKa = 10.28AGYY65 pKa = 8.93GWHH68 pKa = 6.78LNEE71 pKa = 4.7IPLGSSYY78 pKa = 10.73PEE80 pKa = 3.67RR81 pKa = 11.84HH82 pKa = 5.41SDD84 pKa = 3.24KK85 pKa = 11.26VKK87 pKa = 10.02VKK89 pKa = 9.98SFRR92 pKa = 11.84FRR94 pKa = 11.84MQFRR98 pKa = 11.84DD99 pKa = 3.58GANGGTNAANVHH111 pKa = 5.68NLYY114 pKa = 10.72LYY116 pKa = 10.34LVKK119 pKa = 10.73DD120 pKa = 3.66NSGGTEE126 pKa = 3.81VPSFDD131 pKa = 4.81KK132 pKa = 10.72IAMMDD137 pKa = 3.65YY138 pKa = 11.02GNVATAIIDD147 pKa = 3.51HH148 pKa = 7.0DD149 pKa = 4.1SEE151 pKa = 4.28KK152 pKa = 10.68RR153 pKa = 11.84FTIVRR158 pKa = 11.84QWKK161 pKa = 7.72YY162 pKa = 9.11TFTGSCNSQYY172 pKa = 10.15PGKK175 pKa = 10.46DD176 pKa = 2.51RR177 pKa = 11.84IDD179 pKa = 4.13FNDD182 pKa = 3.08VVYY185 pKa = 10.91INVDD189 pKa = 3.68TEE191 pKa = 4.55FKK193 pKa = 10.75SATDD197 pKa = 3.32GKK199 pKa = 10.72YY200 pKa = 11.07ANTQKK205 pKa = 10.77NAWVLYY211 pKa = 9.65IIPQSYY217 pKa = 9.9DD218 pKa = 3.32CVIDD222 pKa = 3.0GHH224 pKa = 5.74VNVRR228 pKa = 11.84FEE230 pKa = 4.3SLVV233 pKa = 3.3
MM1 pKa = 7.38VKK3 pKa = 10.13RR4 pKa = 11.84KK5 pKa = 9.88RR6 pKa = 11.84STGVTTTYY14 pKa = 11.07AKK16 pKa = 10.26RR17 pKa = 11.84SRR19 pKa = 11.84VGSGYY24 pKa = 9.23RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84WSRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84IVRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.55GVYY41 pKa = 9.24ARR43 pKa = 11.84HH44 pKa = 6.21RR45 pKa = 11.84SQSVLHH51 pKa = 6.16SLSNATATKK60 pKa = 10.46GKK62 pKa = 10.28AGYY65 pKa = 8.93GWHH68 pKa = 6.78LNEE71 pKa = 4.7IPLGSSYY78 pKa = 10.73PEE80 pKa = 3.67RR81 pKa = 11.84HH82 pKa = 5.41SDD84 pKa = 3.24KK85 pKa = 11.26VKK87 pKa = 10.02VKK89 pKa = 9.98SFRR92 pKa = 11.84FRR94 pKa = 11.84MQFRR98 pKa = 11.84DD99 pKa = 3.58GANGGTNAANVHH111 pKa = 5.68NLYY114 pKa = 10.72LYY116 pKa = 10.34LVKK119 pKa = 10.73DD120 pKa = 3.66NSGGTEE126 pKa = 3.81VPSFDD131 pKa = 4.81KK132 pKa = 10.72IAMMDD137 pKa = 3.65YY138 pKa = 11.02GNVATAIIDD147 pKa = 3.51HH148 pKa = 7.0DD149 pKa = 4.1SEE151 pKa = 4.28KK152 pKa = 10.68RR153 pKa = 11.84FTIVRR158 pKa = 11.84QWKK161 pKa = 7.72YY162 pKa = 9.11TFTGSCNSQYY172 pKa = 10.15PGKK175 pKa = 10.46DD176 pKa = 2.51RR177 pKa = 11.84IDD179 pKa = 4.13FNDD182 pKa = 3.08VVYY185 pKa = 10.91INVDD189 pKa = 3.68TEE191 pKa = 4.55FKK193 pKa = 10.75SATDD197 pKa = 3.32GKK199 pKa = 10.72YY200 pKa = 11.07ANTQKK205 pKa = 10.77NAWVLYY211 pKa = 9.65IIPQSYY217 pKa = 9.9DD218 pKa = 3.32CVIDD222 pKa = 3.0GHH224 pKa = 5.74VNVRR228 pKa = 11.84FEE230 pKa = 4.3SLVV233 pKa = 3.3
Molecular weight: 26.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1196 |
86 |
335 |
199.3 |
22.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.355 ± 0.756 | 1.589 ± 0.414 |
5.1 ± 0.778 | 5.435 ± 1.016 |
3.261 ± 0.372 | 4.515 ± 0.955 |
2.425 ± 0.394 | 5.1 ± 0.347 |
5.268 ± 0.957 | 8.027 ± 1.104 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.421 ± 0.342 | 4.849 ± 0.409 |
6.522 ± 1.594 | 5.1 ± 0.82 |
7.358 ± 0.865 | 7.776 ± 1.773 |
7.441 ± 1.04 | 5.268 ± 1.007 |
1.756 ± 0.232 | 5.435 ± 0.434 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
