
Wissadula yellow mosaic virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.16
Get precalculated fractions of proteins
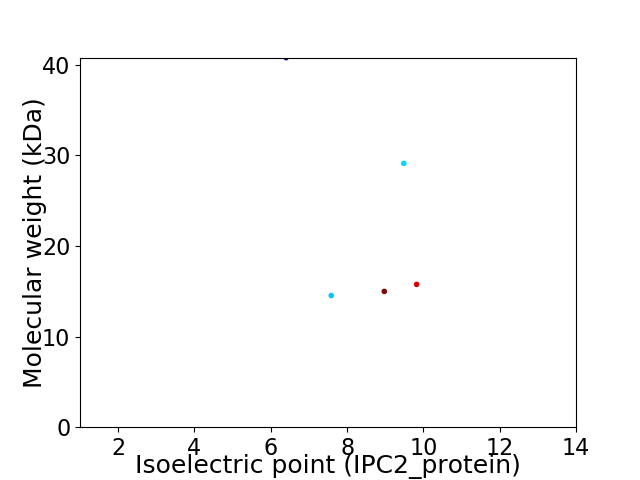
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8GVB5|A0A1D8GVB5_9GEMI Capsid protein OS=Wissadula yellow mosaic virus OX=1904884 GN=CP PE=3 SV=1
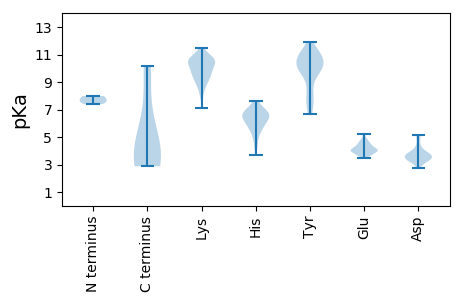
MM1 pKa = 7.39PRR3 pKa = 11.84KK4 pKa = 9.22PSSFRR9 pKa = 11.84LSAKK13 pKa = 10.46NIFLTYY19 pKa = 8.62PQCDD23 pKa = 3.5LSKK26 pKa = 11.49DD27 pKa = 3.33EE28 pKa = 4.72ALQMLHH34 pKa = 5.99ALKK37 pKa = 10.21WSVVKK42 pKa = 8.52PTYY45 pKa = 10.14IRR47 pKa = 11.84VARR50 pKa = 11.84EE51 pKa = 3.67EE52 pKa = 4.28HH53 pKa = 7.09SDD55 pKa = 3.73GLPHH59 pKa = 6.51LHH61 pKa = 7.07CLIQLSGKK69 pKa = 10.24SNIKK73 pKa = 9.95DD74 pKa = 3.22VRR76 pKa = 11.84FFDD79 pKa = 3.57LTHH82 pKa = 6.69PRR84 pKa = 11.84RR85 pKa = 11.84SASFHH90 pKa = 6.84PNVQAAKK97 pKa = 9.12DD98 pKa = 3.63TNAVKK103 pKa = 10.63NYY105 pKa = 6.66ITKK108 pKa = 10.56EE109 pKa = 3.5GDD111 pKa = 3.42YY112 pKa = 10.73CEE114 pKa = 4.54SGQYY118 pKa = 8.42TVSGGTRR125 pKa = 11.84SNKK128 pKa = 10.04DD129 pKa = 3.42DD130 pKa = 3.7VFHH133 pKa = 6.76NAVNAPSVGEE143 pKa = 3.67ALEE146 pKa = 4.46IIKK149 pKa = 10.91AGDD152 pKa = 3.42PKK154 pKa = 10.67TFLVNYY160 pKa = 9.81HH161 pKa = 5.54NLKK164 pKa = 10.84GNLEE168 pKa = 4.05RR169 pKa = 11.84LFAKK173 pKa = 10.39APEE176 pKa = 4.09PWVPPFQLSSFTNVPDD192 pKa = 5.1EE193 pKa = 4.13MQEE196 pKa = 3.8WADD199 pKa = 3.73EE200 pKa = 4.25YY201 pKa = 11.01FGRR204 pKa = 11.84GSAARR209 pKa = 11.84PEE211 pKa = 4.12RR212 pKa = 11.84PISIIVEE219 pKa = 4.05GDD221 pKa = 3.08SRR223 pKa = 11.84TGKK226 pKa = 8.52TMWARR231 pKa = 11.84SLGPHH236 pKa = 6.57NYY238 pKa = 10.17LSGHH242 pKa = 6.86LDD244 pKa = 3.63FNARR248 pKa = 11.84IYY250 pKa = 10.96SNEE253 pKa = 3.69VEE255 pKa = 4.57FNVIDD260 pKa = 4.25DD261 pKa = 4.32VSPQYY266 pKa = 11.29LKK268 pKa = 10.82LKK270 pKa = 9.19HH271 pKa = 5.51WKK273 pKa = 9.67EE274 pKa = 3.98LIGAQKK280 pKa = 10.57DD281 pKa = 3.42WQSNCKK287 pKa = 9.38YY288 pKa = 10.21GKK290 pKa = 9.18PVQIKK295 pKa = 10.76GGMDD299 pKa = 3.62PSNRR303 pKa = 11.84ALQSWSGISYY313 pKa = 11.01KK314 pKa = 10.92DD315 pKa = 3.66FLDD318 pKa = 4.07KK319 pKa = 11.41EE320 pKa = 4.73EE321 pKa = 4.14NTSLRR326 pKa = 11.84NWTIKK331 pKa = 10.32NAVFITLDD339 pKa = 3.31SPLYY343 pKa = 10.31QEE345 pKa = 4.5STQAGQEE352 pKa = 4.07EE353 pKa = 5.23GHH355 pKa = 6.1QEE357 pKa = 3.92TANN360 pKa = 3.42
MM1 pKa = 7.39PRR3 pKa = 11.84KK4 pKa = 9.22PSSFRR9 pKa = 11.84LSAKK13 pKa = 10.46NIFLTYY19 pKa = 8.62PQCDD23 pKa = 3.5LSKK26 pKa = 11.49DD27 pKa = 3.33EE28 pKa = 4.72ALQMLHH34 pKa = 5.99ALKK37 pKa = 10.21WSVVKK42 pKa = 8.52PTYY45 pKa = 10.14IRR47 pKa = 11.84VARR50 pKa = 11.84EE51 pKa = 3.67EE52 pKa = 4.28HH53 pKa = 7.09SDD55 pKa = 3.73GLPHH59 pKa = 6.51LHH61 pKa = 7.07CLIQLSGKK69 pKa = 10.24SNIKK73 pKa = 9.95DD74 pKa = 3.22VRR76 pKa = 11.84FFDD79 pKa = 3.57LTHH82 pKa = 6.69PRR84 pKa = 11.84RR85 pKa = 11.84SASFHH90 pKa = 6.84PNVQAAKK97 pKa = 9.12DD98 pKa = 3.63TNAVKK103 pKa = 10.63NYY105 pKa = 6.66ITKK108 pKa = 10.56EE109 pKa = 3.5GDD111 pKa = 3.42YY112 pKa = 10.73CEE114 pKa = 4.54SGQYY118 pKa = 8.42TVSGGTRR125 pKa = 11.84SNKK128 pKa = 10.04DD129 pKa = 3.42DD130 pKa = 3.7VFHH133 pKa = 6.76NAVNAPSVGEE143 pKa = 3.67ALEE146 pKa = 4.46IIKK149 pKa = 10.91AGDD152 pKa = 3.42PKK154 pKa = 10.67TFLVNYY160 pKa = 9.81HH161 pKa = 5.54NLKK164 pKa = 10.84GNLEE168 pKa = 4.05RR169 pKa = 11.84LFAKK173 pKa = 10.39APEE176 pKa = 4.09PWVPPFQLSSFTNVPDD192 pKa = 5.1EE193 pKa = 4.13MQEE196 pKa = 3.8WADD199 pKa = 3.73EE200 pKa = 4.25YY201 pKa = 11.01FGRR204 pKa = 11.84GSAARR209 pKa = 11.84PEE211 pKa = 4.12RR212 pKa = 11.84PISIIVEE219 pKa = 4.05GDD221 pKa = 3.08SRR223 pKa = 11.84TGKK226 pKa = 8.52TMWARR231 pKa = 11.84SLGPHH236 pKa = 6.57NYY238 pKa = 10.17LSGHH242 pKa = 6.86LDD244 pKa = 3.63FNARR248 pKa = 11.84IYY250 pKa = 10.96SNEE253 pKa = 3.69VEE255 pKa = 4.57FNVIDD260 pKa = 4.25DD261 pKa = 4.32VSPQYY266 pKa = 11.29LKK268 pKa = 10.82LKK270 pKa = 9.19HH271 pKa = 5.51WKK273 pKa = 9.67EE274 pKa = 3.98LIGAQKK280 pKa = 10.57DD281 pKa = 3.42WQSNCKK287 pKa = 9.38YY288 pKa = 10.21GKK290 pKa = 9.18PVQIKK295 pKa = 10.76GGMDD299 pKa = 3.62PSNRR303 pKa = 11.84ALQSWSGISYY313 pKa = 11.01KK314 pKa = 10.92DD315 pKa = 3.66FLDD318 pKa = 4.07KK319 pKa = 11.41EE320 pKa = 4.73EE321 pKa = 4.14NTSLRR326 pKa = 11.84NWTIKK331 pKa = 10.32NAVFITLDD339 pKa = 3.31SPLYY343 pKa = 10.31QEE345 pKa = 4.5STQAGQEE352 pKa = 4.07EE353 pKa = 5.23GHH355 pKa = 6.1QEE357 pKa = 3.92TANN360 pKa = 3.42
Molecular weight: 40.78 kDa
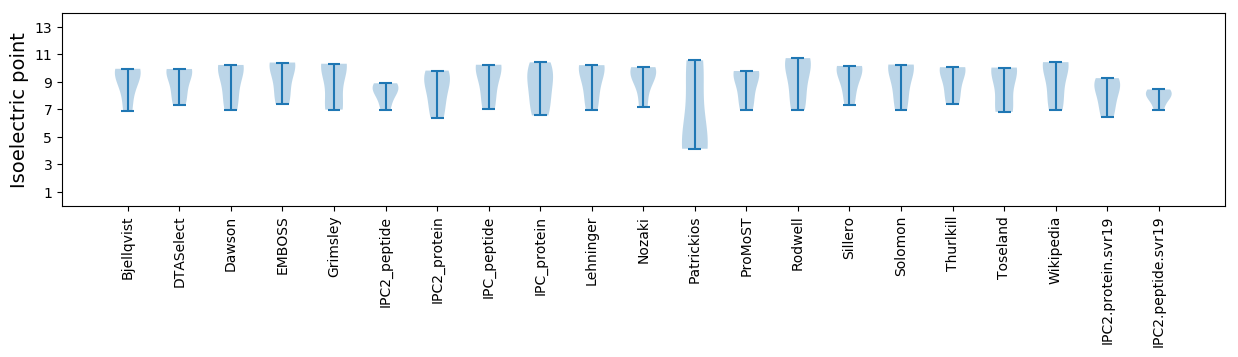
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8GVC6|A0A1D8GVC6_9GEMI Replication enhancer OS=Wissadula yellow mosaic virus OX=1904884 GN=REn PE=3 SV=1
MM1 pKa = 7.98PKK3 pKa = 9.61RR4 pKa = 11.84DD5 pKa = 3.78APWRR9 pKa = 11.84SMAGTSKK16 pKa = 10.56VSRR19 pKa = 11.84NANYY23 pKa = 10.24SPRR26 pKa = 11.84GGYY29 pKa = 8.07GLKK32 pKa = 10.33FNKK35 pKa = 9.46AAEE38 pKa = 4.24WVNRR42 pKa = 11.84PMYY45 pKa = 9.79RR46 pKa = 11.84KK47 pKa = 8.95PRR49 pKa = 11.84IYY51 pKa = 9.6RR52 pKa = 11.84TLRR55 pKa = 11.84TPDD58 pKa = 3.21VPRR61 pKa = 11.84GCEE64 pKa = 4.4GPCKK68 pKa = 9.6IQSYY72 pKa = 7.68DD73 pKa = 3.35QRR75 pKa = 11.84HH76 pKa = 6.66DD77 pKa = 3.18IAHH80 pKa = 6.49TGKK83 pKa = 10.46VMCISDD89 pKa = 3.3VSRR92 pKa = 11.84GNGIAQRR99 pKa = 11.84VGKK102 pKa = 9.34RR103 pKa = 11.84FCVKK107 pKa = 9.73SVYY110 pKa = 10.25IIGKK114 pKa = 8.8VWMDD118 pKa = 4.5DD119 pKa = 3.6NIKK122 pKa = 10.64LKK124 pKa = 10.74NHH126 pKa = 5.99TNSVMFWLVRR136 pKa = 11.84DD137 pKa = 3.75RR138 pKa = 11.84RR139 pKa = 11.84PNGNAMDD146 pKa = 4.77FGHH149 pKa = 7.56LFNMYY154 pKa = 10.64DD155 pKa = 4.01NEE157 pKa = 4.36PSTATVKK164 pKa = 10.85NDD166 pKa = 2.82LRR168 pKa = 11.84DD169 pKa = 3.52RR170 pKa = 11.84FQVLHH175 pKa = 6.7RR176 pKa = 11.84FNAKK180 pKa = 8.04VTGGQYY186 pKa = 11.1ASNEE190 pKa = 3.84QALVRR195 pKa = 11.84RR196 pKa = 11.84FWKK199 pKa = 10.46VNNHH203 pKa = 3.68VTYY206 pKa = 10.85NNQEE210 pKa = 3.68AARR213 pKa = 11.84YY214 pKa = 7.48EE215 pKa = 4.13NHH217 pKa = 6.5TEE219 pKa = 3.94NALLLYY225 pKa = 7.29MACTHH230 pKa = 7.07ASNPVYY236 pKa = 9.86ATLKK240 pKa = 9.47IRR242 pKa = 11.84IYY244 pKa = 10.66FYY246 pKa = 11.34DD247 pKa = 3.3SVLNN251 pKa = 4.02
MM1 pKa = 7.98PKK3 pKa = 9.61RR4 pKa = 11.84DD5 pKa = 3.78APWRR9 pKa = 11.84SMAGTSKK16 pKa = 10.56VSRR19 pKa = 11.84NANYY23 pKa = 10.24SPRR26 pKa = 11.84GGYY29 pKa = 8.07GLKK32 pKa = 10.33FNKK35 pKa = 9.46AAEE38 pKa = 4.24WVNRR42 pKa = 11.84PMYY45 pKa = 9.79RR46 pKa = 11.84KK47 pKa = 8.95PRR49 pKa = 11.84IYY51 pKa = 9.6RR52 pKa = 11.84TLRR55 pKa = 11.84TPDD58 pKa = 3.21VPRR61 pKa = 11.84GCEE64 pKa = 4.4GPCKK68 pKa = 9.6IQSYY72 pKa = 7.68DD73 pKa = 3.35QRR75 pKa = 11.84HH76 pKa = 6.66DD77 pKa = 3.18IAHH80 pKa = 6.49TGKK83 pKa = 10.46VMCISDD89 pKa = 3.3VSRR92 pKa = 11.84GNGIAQRR99 pKa = 11.84VGKK102 pKa = 9.34RR103 pKa = 11.84FCVKK107 pKa = 9.73SVYY110 pKa = 10.25IIGKK114 pKa = 8.8VWMDD118 pKa = 4.5DD119 pKa = 3.6NIKK122 pKa = 10.64LKK124 pKa = 10.74NHH126 pKa = 5.99TNSVMFWLVRR136 pKa = 11.84DD137 pKa = 3.75RR138 pKa = 11.84RR139 pKa = 11.84PNGNAMDD146 pKa = 4.77FGHH149 pKa = 7.56LFNMYY154 pKa = 10.64DD155 pKa = 4.01NEE157 pKa = 4.36PSTATVKK164 pKa = 10.85NDD166 pKa = 2.82LRR168 pKa = 11.84DD169 pKa = 3.52RR170 pKa = 11.84FQVLHH175 pKa = 6.7RR176 pKa = 11.84FNAKK180 pKa = 8.04VTGGQYY186 pKa = 11.1ASNEE190 pKa = 3.84QALVRR195 pKa = 11.84RR196 pKa = 11.84FWKK199 pKa = 10.46VNNHH203 pKa = 3.68VTYY206 pKa = 10.85NNQEE210 pKa = 3.68AARR213 pKa = 11.84YY214 pKa = 7.48EE215 pKa = 4.13NHH217 pKa = 6.5TEE219 pKa = 3.94NALLLYY225 pKa = 7.29MACTHH230 pKa = 7.07ASNPVYY236 pKa = 9.86ATLKK240 pKa = 9.47IRR242 pKa = 11.84IYY244 pKa = 10.66FYY246 pKa = 11.34DD247 pKa = 3.3SVLNN251 pKa = 4.02
Molecular weight: 29.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1003 |
129 |
360 |
200.6 |
23.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.484 ± 0.927 | 1.695 ± 0.588 |
4.586 ± 0.673 | 4.686 ± 0.765 |
4.088 ± 0.245 | 5.184 ± 0.64 |
3.689 ± 0.56 | 5.085 ± 0.494 |
5.882 ± 0.766 | 7.178 ± 0.738 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.193 ± 0.604 | 6.68 ± 0.756 |
5.384 ± 0.604 | 4.586 ± 0.956 |
6.879 ± 1.001 | 8.275 ± 1.325 |
6.281 ± 1.208 | 6.082 ± 0.898 |
1.894 ± 0.168 | 4.187 ± 0.714 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
