
Lake Sarah-associated circular molecule 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 9.19
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G993|A0A126G993_9VIRU Replication associated protein OS=Lake Sarah-associated circular molecule 1 OX=1685724 PE=4 SV=1
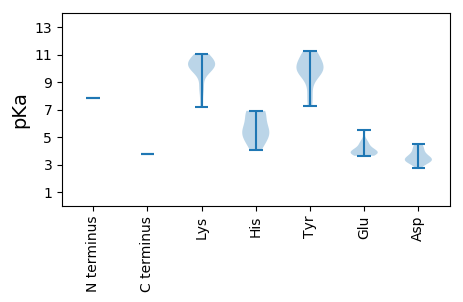
MM1 pKa = 7.81ADD3 pKa = 3.31YY4 pKa = 10.86AGKK7 pKa = 7.45TKK9 pKa = 10.8YY10 pKa = 9.71WIFVLKK16 pKa = 9.83TRR18 pKa = 11.84DD19 pKa = 3.54PARR22 pKa = 11.84SSIPIKK28 pKa = 10.22RR29 pKa = 11.84ATRR32 pKa = 11.84PTYY35 pKa = 10.21ATDD38 pKa = 3.22TKK40 pKa = 10.26SASRR44 pKa = 11.84ARR46 pKa = 11.84PHH48 pKa = 5.38LQGYY52 pKa = 9.79IEE54 pKa = 4.55MINRR58 pKa = 11.84KK59 pKa = 7.18TMIQMKK65 pKa = 10.1HH66 pKa = 6.74DD67 pKa = 3.68INPRR71 pKa = 11.84CKK73 pKa = 8.59WLPRR77 pKa = 11.84GGTALEE83 pKa = 3.91AATYY87 pKa = 9.97AKK89 pKa = 10.3KK90 pKa = 10.72DD91 pKa = 3.29NTGIYY96 pKa = 9.93EE97 pKa = 4.53DD98 pKa = 4.39GVLSKK103 pKa = 10.82SDD105 pKa = 3.13QGKK108 pKa = 9.71RR109 pKa = 11.84NDD111 pKa = 3.25MAEE114 pKa = 3.82IFKK117 pKa = 10.12MIEE120 pKa = 3.81NGADD124 pKa = 3.2EE125 pKa = 5.5ADD127 pKa = 3.34ICRR130 pKa = 11.84KK131 pKa = 10.14YY132 pKa = 9.66PGQFIRR138 pKa = 11.84YY139 pKa = 7.83HH140 pKa = 6.91AGITKK145 pKa = 10.33AISLQFQKK153 pKa = 10.65RR154 pKa = 11.84VGDD157 pKa = 3.43IEE159 pKa = 3.93IYY161 pKa = 10.45YY162 pKa = 9.89IYY164 pKa = 10.74GKK166 pKa = 8.41TGSGKK171 pKa = 8.62TSSVVKK177 pKa = 10.77KK178 pKa = 10.31EE179 pKa = 3.89GEE181 pKa = 4.25ALYY184 pKa = 8.55HH185 pKa = 6.78TYY187 pKa = 10.85DD188 pKa = 3.68QDD190 pKa = 4.19LRR192 pKa = 11.84WFDD195 pKa = 3.49GYY197 pKa = 11.18KK198 pKa = 10.05PSRR201 pKa = 11.84HH202 pKa = 5.0SAILIDD208 pKa = 4.51EE209 pKa = 4.68FNGSQKK215 pKa = 11.0AVFYY219 pKa = 11.27NKK221 pKa = 9.85ILDD224 pKa = 4.1RR225 pKa = 11.84YY226 pKa = 7.24TPWVEE231 pKa = 3.85VKK233 pKa = 10.13GAKK236 pKa = 9.63LRR238 pKa = 11.84ITAKK242 pKa = 10.3RR243 pKa = 11.84IYY245 pKa = 9.93ICSNYY250 pKa = 9.69KK251 pKa = 10.21LKK253 pKa = 10.77EE254 pKa = 3.9VANTQGWSPEE264 pKa = 3.64EE265 pKa = 3.74LEE267 pKa = 4.06QFKK270 pKa = 11.04RR271 pKa = 11.84RR272 pKa = 11.84VTKK275 pKa = 10.35FIHH278 pKa = 5.59VEE280 pKa = 4.02RR281 pKa = 11.84LPGAPPVDD289 pKa = 2.77IWAPLVPQAAEE300 pKa = 3.73PTTPVRR306 pKa = 11.84PQHH309 pKa = 4.77PHH311 pKa = 4.07GWRR314 pKa = 11.84QRR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84ATPPTCADD326 pKa = 3.91SACSGAPAGSRR337 pKa = 11.84HH338 pKa = 5.61SMRR341 pKa = 11.84VTNAPRR347 pKa = 11.84SGILCSRR354 pKa = 11.84SRR356 pKa = 11.84NRR358 pKa = 3.75
MM1 pKa = 7.81ADD3 pKa = 3.31YY4 pKa = 10.86AGKK7 pKa = 7.45TKK9 pKa = 10.8YY10 pKa = 9.71WIFVLKK16 pKa = 9.83TRR18 pKa = 11.84DD19 pKa = 3.54PARR22 pKa = 11.84SSIPIKK28 pKa = 10.22RR29 pKa = 11.84ATRR32 pKa = 11.84PTYY35 pKa = 10.21ATDD38 pKa = 3.22TKK40 pKa = 10.26SASRR44 pKa = 11.84ARR46 pKa = 11.84PHH48 pKa = 5.38LQGYY52 pKa = 9.79IEE54 pKa = 4.55MINRR58 pKa = 11.84KK59 pKa = 7.18TMIQMKK65 pKa = 10.1HH66 pKa = 6.74DD67 pKa = 3.68INPRR71 pKa = 11.84CKK73 pKa = 8.59WLPRR77 pKa = 11.84GGTALEE83 pKa = 3.91AATYY87 pKa = 9.97AKK89 pKa = 10.3KK90 pKa = 10.72DD91 pKa = 3.29NTGIYY96 pKa = 9.93EE97 pKa = 4.53DD98 pKa = 4.39GVLSKK103 pKa = 10.82SDD105 pKa = 3.13QGKK108 pKa = 9.71RR109 pKa = 11.84NDD111 pKa = 3.25MAEE114 pKa = 3.82IFKK117 pKa = 10.12MIEE120 pKa = 3.81NGADD124 pKa = 3.2EE125 pKa = 5.5ADD127 pKa = 3.34ICRR130 pKa = 11.84KK131 pKa = 10.14YY132 pKa = 9.66PGQFIRR138 pKa = 11.84YY139 pKa = 7.83HH140 pKa = 6.91AGITKK145 pKa = 10.33AISLQFQKK153 pKa = 10.65RR154 pKa = 11.84VGDD157 pKa = 3.43IEE159 pKa = 3.93IYY161 pKa = 10.45YY162 pKa = 9.89IYY164 pKa = 10.74GKK166 pKa = 8.41TGSGKK171 pKa = 8.62TSSVVKK177 pKa = 10.77KK178 pKa = 10.31EE179 pKa = 3.89GEE181 pKa = 4.25ALYY184 pKa = 8.55HH185 pKa = 6.78TYY187 pKa = 10.85DD188 pKa = 3.68QDD190 pKa = 4.19LRR192 pKa = 11.84WFDD195 pKa = 3.49GYY197 pKa = 11.18KK198 pKa = 10.05PSRR201 pKa = 11.84HH202 pKa = 5.0SAILIDD208 pKa = 4.51EE209 pKa = 4.68FNGSQKK215 pKa = 11.0AVFYY219 pKa = 11.27NKK221 pKa = 9.85ILDD224 pKa = 4.1RR225 pKa = 11.84YY226 pKa = 7.24TPWVEE231 pKa = 3.85VKK233 pKa = 10.13GAKK236 pKa = 9.63LRR238 pKa = 11.84ITAKK242 pKa = 10.3RR243 pKa = 11.84IYY245 pKa = 9.93ICSNYY250 pKa = 9.69KK251 pKa = 10.21LKK253 pKa = 10.77EE254 pKa = 3.9VANTQGWSPEE264 pKa = 3.64EE265 pKa = 3.74LEE267 pKa = 4.06QFKK270 pKa = 11.04RR271 pKa = 11.84RR272 pKa = 11.84VTKK275 pKa = 10.35FIHH278 pKa = 5.59VEE280 pKa = 4.02RR281 pKa = 11.84LPGAPPVDD289 pKa = 2.77IWAPLVPQAAEE300 pKa = 3.73PTTPVRR306 pKa = 11.84PQHH309 pKa = 4.77PHH311 pKa = 4.07GWRR314 pKa = 11.84QRR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84ATPPTCADD326 pKa = 3.91SACSGAPAGSRR337 pKa = 11.84HH338 pKa = 5.61SMRR341 pKa = 11.84VTNAPRR347 pKa = 11.84SGILCSRR354 pKa = 11.84SRR356 pKa = 11.84NRR358 pKa = 3.75
Molecular weight: 40.74 kDa
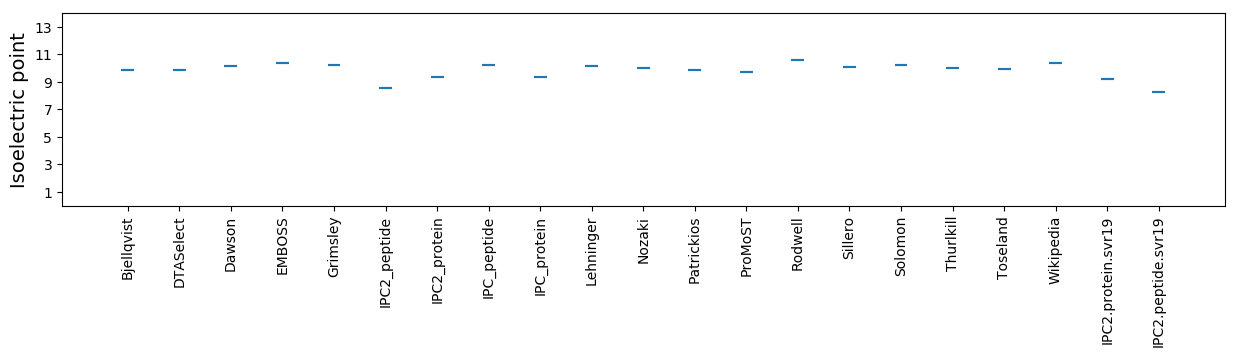
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G993|A0A126G993_9VIRU Replication associated protein OS=Lake Sarah-associated circular molecule 1 OX=1685724 PE=4 SV=1
MM1 pKa = 7.81ADD3 pKa = 3.31YY4 pKa = 10.86AGKK7 pKa = 7.45TKK9 pKa = 10.8YY10 pKa = 9.71WIFVLKK16 pKa = 9.83TRR18 pKa = 11.84DD19 pKa = 3.54PARR22 pKa = 11.84SSIPIKK28 pKa = 10.22RR29 pKa = 11.84ATRR32 pKa = 11.84PTYY35 pKa = 10.21ATDD38 pKa = 3.22TKK40 pKa = 10.26SASRR44 pKa = 11.84ARR46 pKa = 11.84PHH48 pKa = 5.38LQGYY52 pKa = 9.79IEE54 pKa = 4.55MINRR58 pKa = 11.84KK59 pKa = 7.18TMIQMKK65 pKa = 10.1HH66 pKa = 6.74DD67 pKa = 3.68INPRR71 pKa = 11.84CKK73 pKa = 8.59WLPRR77 pKa = 11.84GGTALEE83 pKa = 3.91AATYY87 pKa = 9.97AKK89 pKa = 10.3KK90 pKa = 10.72DD91 pKa = 3.29NTGIYY96 pKa = 9.93EE97 pKa = 4.53DD98 pKa = 4.39GVLSKK103 pKa = 10.82SDD105 pKa = 3.13QGKK108 pKa = 9.71RR109 pKa = 11.84NDD111 pKa = 3.25MAEE114 pKa = 3.82IFKK117 pKa = 10.12MIEE120 pKa = 3.81NGADD124 pKa = 3.2EE125 pKa = 5.5ADD127 pKa = 3.34ICRR130 pKa = 11.84KK131 pKa = 10.14YY132 pKa = 9.66PGQFIRR138 pKa = 11.84YY139 pKa = 7.83HH140 pKa = 6.91AGITKK145 pKa = 10.33AISLQFQKK153 pKa = 10.65RR154 pKa = 11.84VGDD157 pKa = 3.43IEE159 pKa = 3.93IYY161 pKa = 10.45YY162 pKa = 9.89IYY164 pKa = 10.74GKK166 pKa = 8.41TGSGKK171 pKa = 8.62TSSVVKK177 pKa = 10.77KK178 pKa = 10.31EE179 pKa = 3.89GEE181 pKa = 4.25ALYY184 pKa = 8.55HH185 pKa = 6.78TYY187 pKa = 10.85DD188 pKa = 3.68QDD190 pKa = 4.19LRR192 pKa = 11.84WFDD195 pKa = 3.49GYY197 pKa = 11.18KK198 pKa = 10.05PSRR201 pKa = 11.84HH202 pKa = 5.0SAILIDD208 pKa = 4.51EE209 pKa = 4.68FNGSQKK215 pKa = 11.0AVFYY219 pKa = 11.27NKK221 pKa = 9.85ILDD224 pKa = 4.1RR225 pKa = 11.84YY226 pKa = 7.24TPWVEE231 pKa = 3.85VKK233 pKa = 10.13GAKK236 pKa = 9.63LRR238 pKa = 11.84ITAKK242 pKa = 10.3RR243 pKa = 11.84IYY245 pKa = 9.93ICSNYY250 pKa = 9.69KK251 pKa = 10.21LKK253 pKa = 10.77EE254 pKa = 3.9VANTQGWSPEE264 pKa = 3.64EE265 pKa = 3.74LEE267 pKa = 4.06QFKK270 pKa = 11.04RR271 pKa = 11.84RR272 pKa = 11.84VTKK275 pKa = 10.35FIHH278 pKa = 5.59VEE280 pKa = 4.02RR281 pKa = 11.84LPGAPPVDD289 pKa = 2.77IWAPLVPQAAEE300 pKa = 3.73PTTPVRR306 pKa = 11.84PQHH309 pKa = 4.77PHH311 pKa = 4.07GWRR314 pKa = 11.84QRR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84ATPPTCADD326 pKa = 3.91SACSGAPAGSRR337 pKa = 11.84HH338 pKa = 5.61SMRR341 pKa = 11.84VTNAPRR347 pKa = 11.84SGILCSRR354 pKa = 11.84SRR356 pKa = 11.84NRR358 pKa = 3.75
MM1 pKa = 7.81ADD3 pKa = 3.31YY4 pKa = 10.86AGKK7 pKa = 7.45TKK9 pKa = 10.8YY10 pKa = 9.71WIFVLKK16 pKa = 9.83TRR18 pKa = 11.84DD19 pKa = 3.54PARR22 pKa = 11.84SSIPIKK28 pKa = 10.22RR29 pKa = 11.84ATRR32 pKa = 11.84PTYY35 pKa = 10.21ATDD38 pKa = 3.22TKK40 pKa = 10.26SASRR44 pKa = 11.84ARR46 pKa = 11.84PHH48 pKa = 5.38LQGYY52 pKa = 9.79IEE54 pKa = 4.55MINRR58 pKa = 11.84KK59 pKa = 7.18TMIQMKK65 pKa = 10.1HH66 pKa = 6.74DD67 pKa = 3.68INPRR71 pKa = 11.84CKK73 pKa = 8.59WLPRR77 pKa = 11.84GGTALEE83 pKa = 3.91AATYY87 pKa = 9.97AKK89 pKa = 10.3KK90 pKa = 10.72DD91 pKa = 3.29NTGIYY96 pKa = 9.93EE97 pKa = 4.53DD98 pKa = 4.39GVLSKK103 pKa = 10.82SDD105 pKa = 3.13QGKK108 pKa = 9.71RR109 pKa = 11.84NDD111 pKa = 3.25MAEE114 pKa = 3.82IFKK117 pKa = 10.12MIEE120 pKa = 3.81NGADD124 pKa = 3.2EE125 pKa = 5.5ADD127 pKa = 3.34ICRR130 pKa = 11.84KK131 pKa = 10.14YY132 pKa = 9.66PGQFIRR138 pKa = 11.84YY139 pKa = 7.83HH140 pKa = 6.91AGITKK145 pKa = 10.33AISLQFQKK153 pKa = 10.65RR154 pKa = 11.84VGDD157 pKa = 3.43IEE159 pKa = 3.93IYY161 pKa = 10.45YY162 pKa = 9.89IYY164 pKa = 10.74GKK166 pKa = 8.41TGSGKK171 pKa = 8.62TSSVVKK177 pKa = 10.77KK178 pKa = 10.31EE179 pKa = 3.89GEE181 pKa = 4.25ALYY184 pKa = 8.55HH185 pKa = 6.78TYY187 pKa = 10.85DD188 pKa = 3.68QDD190 pKa = 4.19LRR192 pKa = 11.84WFDD195 pKa = 3.49GYY197 pKa = 11.18KK198 pKa = 10.05PSRR201 pKa = 11.84HH202 pKa = 5.0SAILIDD208 pKa = 4.51EE209 pKa = 4.68FNGSQKK215 pKa = 11.0AVFYY219 pKa = 11.27NKK221 pKa = 9.85ILDD224 pKa = 4.1RR225 pKa = 11.84YY226 pKa = 7.24TPWVEE231 pKa = 3.85VKK233 pKa = 10.13GAKK236 pKa = 9.63LRR238 pKa = 11.84ITAKK242 pKa = 10.3RR243 pKa = 11.84IYY245 pKa = 9.93ICSNYY250 pKa = 9.69KK251 pKa = 10.21LKK253 pKa = 10.77EE254 pKa = 3.9VANTQGWSPEE264 pKa = 3.64EE265 pKa = 3.74LEE267 pKa = 4.06QFKK270 pKa = 11.04RR271 pKa = 11.84RR272 pKa = 11.84VTKK275 pKa = 10.35FIHH278 pKa = 5.59VEE280 pKa = 4.02RR281 pKa = 11.84LPGAPPVDD289 pKa = 2.77IWAPLVPQAAEE300 pKa = 3.73PTTPVRR306 pKa = 11.84PQHH309 pKa = 4.77PHH311 pKa = 4.07GWRR314 pKa = 11.84QRR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84ATPPTCADD326 pKa = 3.91SACSGAPAGSRR337 pKa = 11.84HH338 pKa = 5.61SMRR341 pKa = 11.84VTNAPRR347 pKa = 11.84SGILCSRR354 pKa = 11.84SRR356 pKa = 11.84NRR358 pKa = 3.75
Molecular weight: 40.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
358 |
358 |
358 |
358.0 |
40.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.939 ± 0.0 | 1.676 ± 0.0 |
5.028 ± 0.0 | 4.749 ± 0.0 |
2.514 ± 0.0 | 6.704 ± 0.0 |
2.514 ± 0.0 | 7.263 ± 0.0 |
8.38 ± 0.0 | 4.469 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.955 ± 0.0 | 3.073 ± 0.0 |
6.425 ± 0.0 | 3.631 ± 0.0 |
8.939 ± 0.0 | 6.145 ± 0.0 |
6.425 ± 0.0 | 4.19 ± 0.0 |
1.955 ± 0.0 | 5.028 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
