
Polaribacter gangjinensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
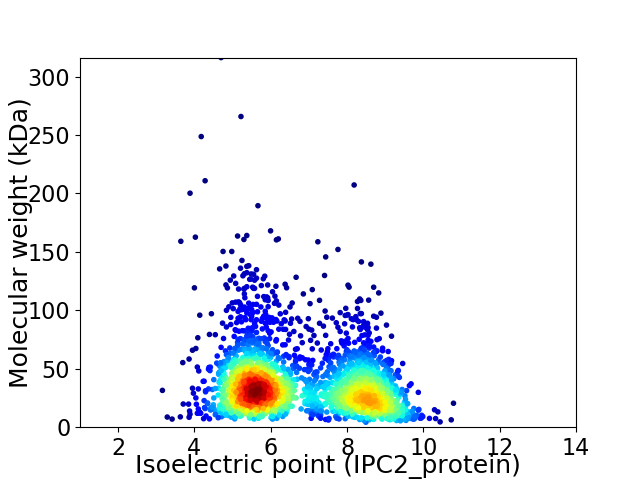
Virtual 2D-PAGE plot for 2512 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
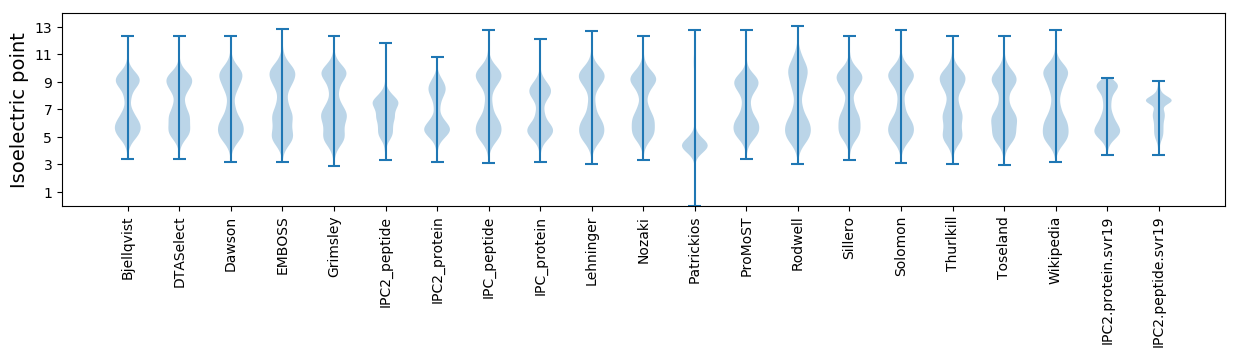
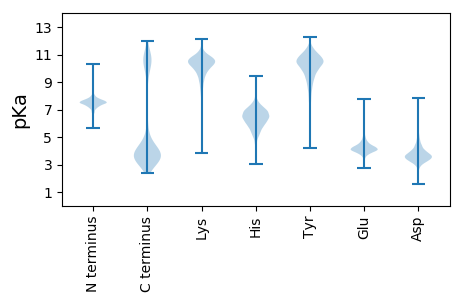
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7WC78|A0A2S7WC78_9FLAO Two-component sensor histidine kinase OS=Polaribacter gangjinensis OX=574710 GN=BTO13_08210 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.39KK3 pKa = 10.39NYY5 pKa = 9.54FLSLFTLLFITVSAFAQDD23 pKa = 3.68PVINEE28 pKa = 4.57LDD30 pKa = 3.7ADD32 pKa = 4.22TPGTDD37 pKa = 2.77TAEE40 pKa = 4.5FIEE43 pKa = 5.14IKK45 pKa = 8.13WTPNTSLTGYY55 pKa = 10.06IVVLYY60 pKa = 10.51NGSNDD65 pKa = 3.13LSYY68 pKa = 10.73YY69 pKa = 10.54TIDD72 pKa = 4.16LSGKK76 pKa = 8.39STDD79 pKa = 3.28ANGFFMIASAGLASGDD95 pKa = 4.88DD96 pKa = 3.44IVIPDD101 pKa = 3.5NTLQNGADD109 pKa = 3.54AVALYY114 pKa = 9.77QAAAGDD120 pKa = 4.37FPNGTAVTSTNLIDD134 pKa = 4.41ALIYY138 pKa = 8.11DD139 pKa = 4.64TNDD142 pKa = 2.9QDD144 pKa = 5.33DD145 pKa = 4.22AGLLSGLGEE154 pKa = 3.97TVQYY158 pKa = 11.34NEE160 pKa = 5.06DD161 pKa = 3.44GGGNKK166 pKa = 9.78DD167 pKa = 3.05NHH169 pKa = 6.51SIQRR173 pKa = 11.84KK174 pKa = 9.58DD175 pKa = 3.37DD176 pKa = 3.18GTYY179 pKa = 10.58AVAIPTYY186 pKa = 9.67RR187 pKa = 11.84AEE189 pKa = 4.35NVFGDD194 pKa = 3.94PCVLNLTTINTTCDD208 pKa = 3.15ANTNGVDD215 pKa = 3.55TYY217 pKa = 9.32TTTIDD222 pKa = 3.56FTGGNTGITYY232 pKa = 8.32TITAKK237 pKa = 10.59DD238 pKa = 3.82GSSADD243 pKa = 3.53VGSIGGDD250 pKa = 3.08NPNSVEE256 pKa = 4.19SGTIIITGVNEE267 pKa = 4.19VVNFTVNVKK276 pKa = 10.34GGAGSSCDD284 pKa = 3.01IDD286 pKa = 4.73RR287 pKa = 11.84SINAPTCLPAATCPAQGAVIITEE310 pKa = 4.22IMQNPNFVGDD320 pKa = 3.69DD321 pKa = 3.13AGEE324 pKa = 3.97YY325 pKa = 10.46FEE327 pKa = 6.23IYY329 pKa = 8.82NTTNTPIDD337 pKa = 3.73LKK339 pKa = 11.13GWIIKK344 pKa = 7.74TASTGSPVEE353 pKa = 4.04HH354 pKa = 6.89VIPTSLILPANGYY367 pKa = 9.69LVLGEE372 pKa = 4.17NADD375 pKa = 3.62INSNGGVTVDD385 pKa = 3.71YY386 pKa = 10.8QYY388 pKa = 11.85DD389 pKa = 3.42AALFLGNSTSTVTIEE404 pKa = 4.25CSQSVFDD411 pKa = 4.22TVTYY415 pKa = 10.9DD416 pKa = 4.58DD417 pKa = 5.0GATFPDD423 pKa = 4.03PSGASMEE430 pKa = 4.3LATNKK435 pKa = 9.98YY436 pKa = 10.33SHH438 pKa = 7.13TDD440 pKa = 3.02NDD442 pKa = 4.07NGINWATATAEE453 pKa = 4.44IITGGDD459 pKa = 2.85KK460 pKa = 9.86GTPGAANSFVLSVIKK475 pKa = 10.89NNIIEE480 pKa = 4.44GFATYY485 pKa = 9.1PNPVISDD492 pKa = 3.48NFTITSNSADD502 pKa = 3.5KK503 pKa = 11.38KK504 pKa = 10.3EE505 pKa = 3.91VTIFNVLGKK514 pKa = 10.11KK515 pKa = 9.77VLSTTFSGTKK525 pKa = 9.96SDD527 pKa = 3.74INVATISSGIYY538 pKa = 8.74ILKK541 pKa = 10.15VSEE544 pKa = 4.17GSKK547 pKa = 10.01MSTSKK552 pKa = 11.17LIIKK556 pKa = 9.74
MM1 pKa = 7.48KK2 pKa = 10.39KK3 pKa = 10.39NYY5 pKa = 9.54FLSLFTLLFITVSAFAQDD23 pKa = 3.68PVINEE28 pKa = 4.57LDD30 pKa = 3.7ADD32 pKa = 4.22TPGTDD37 pKa = 2.77TAEE40 pKa = 4.5FIEE43 pKa = 5.14IKK45 pKa = 8.13WTPNTSLTGYY55 pKa = 10.06IVVLYY60 pKa = 10.51NGSNDD65 pKa = 3.13LSYY68 pKa = 10.73YY69 pKa = 10.54TIDD72 pKa = 4.16LSGKK76 pKa = 8.39STDD79 pKa = 3.28ANGFFMIASAGLASGDD95 pKa = 4.88DD96 pKa = 3.44IVIPDD101 pKa = 3.5NTLQNGADD109 pKa = 3.54AVALYY114 pKa = 9.77QAAAGDD120 pKa = 4.37FPNGTAVTSTNLIDD134 pKa = 4.41ALIYY138 pKa = 8.11DD139 pKa = 4.64TNDD142 pKa = 2.9QDD144 pKa = 5.33DD145 pKa = 4.22AGLLSGLGEE154 pKa = 3.97TVQYY158 pKa = 11.34NEE160 pKa = 5.06DD161 pKa = 3.44GGGNKK166 pKa = 9.78DD167 pKa = 3.05NHH169 pKa = 6.51SIQRR173 pKa = 11.84KK174 pKa = 9.58DD175 pKa = 3.37DD176 pKa = 3.18GTYY179 pKa = 10.58AVAIPTYY186 pKa = 9.67RR187 pKa = 11.84AEE189 pKa = 4.35NVFGDD194 pKa = 3.94PCVLNLTTINTTCDD208 pKa = 3.15ANTNGVDD215 pKa = 3.55TYY217 pKa = 9.32TTTIDD222 pKa = 3.56FTGGNTGITYY232 pKa = 8.32TITAKK237 pKa = 10.59DD238 pKa = 3.82GSSADD243 pKa = 3.53VGSIGGDD250 pKa = 3.08NPNSVEE256 pKa = 4.19SGTIIITGVNEE267 pKa = 4.19VVNFTVNVKK276 pKa = 10.34GGAGSSCDD284 pKa = 3.01IDD286 pKa = 4.73RR287 pKa = 11.84SINAPTCLPAATCPAQGAVIITEE310 pKa = 4.22IMQNPNFVGDD320 pKa = 3.69DD321 pKa = 3.13AGEE324 pKa = 3.97YY325 pKa = 10.46FEE327 pKa = 6.23IYY329 pKa = 8.82NTTNTPIDD337 pKa = 3.73LKK339 pKa = 11.13GWIIKK344 pKa = 7.74TASTGSPVEE353 pKa = 4.04HH354 pKa = 6.89VIPTSLILPANGYY367 pKa = 9.69LVLGEE372 pKa = 4.17NADD375 pKa = 3.62INSNGGVTVDD385 pKa = 3.71YY386 pKa = 10.8QYY388 pKa = 11.85DD389 pKa = 3.42AALFLGNSTSTVTIEE404 pKa = 4.25CSQSVFDD411 pKa = 4.22TVTYY415 pKa = 10.9DD416 pKa = 4.58DD417 pKa = 5.0GATFPDD423 pKa = 4.03PSGASMEE430 pKa = 4.3LATNKK435 pKa = 9.98YY436 pKa = 10.33SHH438 pKa = 7.13TDD440 pKa = 3.02NDD442 pKa = 4.07NGINWATATAEE453 pKa = 4.44IITGGDD459 pKa = 2.85KK460 pKa = 9.86GTPGAANSFVLSVIKK475 pKa = 10.89NNIIEE480 pKa = 4.44GFATYY485 pKa = 9.1PNPVISDD492 pKa = 3.48NFTITSNSADD502 pKa = 3.5KK503 pKa = 11.38KK504 pKa = 10.3EE505 pKa = 3.91VTIFNVLGKK514 pKa = 10.11KK515 pKa = 9.77VLSTTFSGTKK525 pKa = 9.96SDD527 pKa = 3.74INVATISSGIYY538 pKa = 8.74ILKK541 pKa = 10.15VSEE544 pKa = 4.17GSKK547 pKa = 10.01MSTSKK552 pKa = 11.17LIIKK556 pKa = 9.74
Molecular weight: 58.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7WBL6|A0A2S7WBL6_9FLAO Malate dehydrogenase OS=Polaribacter gangjinensis OX=574710 GN=mdh PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.6KK28 pKa = 10.83PEE30 pKa = 4.14KK31 pKa = 10.66SLISSKK37 pKa = 10.75KK38 pKa = 7.9STGGRR43 pKa = 11.84NSQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.61KK59 pKa = 9.66QKK61 pKa = 10.66YY62 pKa = 8.25RR63 pKa = 11.84AIDD66 pKa = 3.77FKK68 pKa = 10.9RR69 pKa = 11.84DD70 pKa = 3.24KK71 pKa = 10.39TGIPATVKK79 pKa = 10.04TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALLFYY96 pKa = 11.17ADD98 pKa = 3.45GEE100 pKa = 4.23KK101 pKa = 10.28RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLEE111 pKa = 4.0VDD113 pKa = 3.61QVIISGSGIAPEE125 pKa = 4.4IGNTLPLSEE134 pKa = 4.99IPLGTTISCIEE145 pKa = 4.03LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.1TRR182 pKa = 11.84LVLLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 8.88AGRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.28GYY255 pKa = 7.21KK256 pKa = 8.8TRR258 pKa = 11.84SKK260 pKa = 9.63TKK262 pKa = 10.14ASNKK266 pKa = 9.84YY267 pKa = 8.91IVEE270 pKa = 4.08RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.6KK28 pKa = 10.83PEE30 pKa = 4.14KK31 pKa = 10.66SLISSKK37 pKa = 10.75KK38 pKa = 7.9STGGRR43 pKa = 11.84NSQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.61KK59 pKa = 9.66QKK61 pKa = 10.66YY62 pKa = 8.25RR63 pKa = 11.84AIDD66 pKa = 3.77FKK68 pKa = 10.9RR69 pKa = 11.84DD70 pKa = 3.24KK71 pKa = 10.39TGIPATVKK79 pKa = 10.04TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALLFYY96 pKa = 11.17ADD98 pKa = 3.45GEE100 pKa = 4.23KK101 pKa = 10.28RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLEE111 pKa = 4.0VDD113 pKa = 3.61QVIISGSGIAPEE125 pKa = 4.4IGNTLPLSEE134 pKa = 4.99IPLGTTISCIEE145 pKa = 4.03LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.1TRR182 pKa = 11.84LVLLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 8.88AGRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.28GYY255 pKa = 7.21KK256 pKa = 8.8TRR258 pKa = 11.84SKK260 pKa = 9.63TKK262 pKa = 10.14ASNKK266 pKa = 9.84YY267 pKa = 8.91IVEE270 pKa = 4.08RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
Molecular weight: 29.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
883914 |
38 |
2858 |
351.9 |
39.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.206 ± 0.048 | 0.675 ± 0.014 |
5.225 ± 0.029 | 6.549 ± 0.05 |
5.806 ± 0.048 | 6.122 ± 0.047 |
1.704 ± 0.02 | 8.615 ± 0.042 |
8.349 ± 0.064 | 9.045 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.087 ± 0.024 | 6.622 ± 0.054 |
3.243 ± 0.026 | 3.527 ± 0.028 |
3.072 ± 0.032 | 6.517 ± 0.042 |
5.864 ± 0.051 | 5.979 ± 0.036 |
1.012 ± 0.018 | 3.779 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
