
Youcai mosaic virus (YoMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
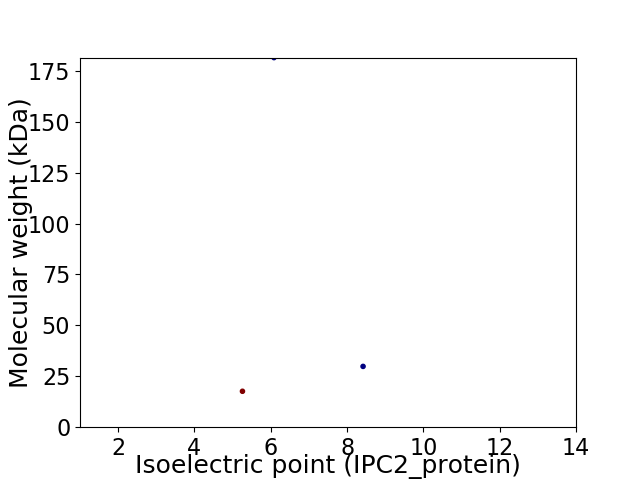
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
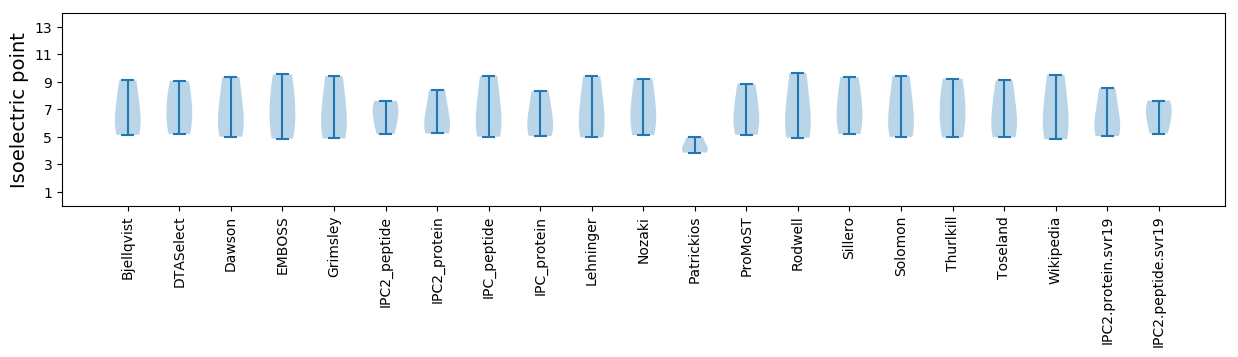
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q66222|CAPSD_YOMV Capsid protein OS=Youcai mosaic virus OX=228578 GN=CP PE=2 SV=3
MM1 pKa = 7.52VYY3 pKa = 10.67NITSSNQYY11 pKa = 10.63QYY13 pKa = 10.8FAAMWAEE20 pKa = 4.06PTAMLNQCVSALSQSYY36 pKa = 7.22QTQAARR42 pKa = 11.84DD43 pKa = 3.67TVRR46 pKa = 11.84QQFSNLLSAIVTPNQRR62 pKa = 11.84FPEE65 pKa = 4.32AGYY68 pKa = 10.16RR69 pKa = 11.84VYY71 pKa = 10.85INSAVLKK78 pKa = 9.44PLYY81 pKa = 9.69EE82 pKa = 4.29SLMKK86 pKa = 10.92SFDD89 pKa = 3.23TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.26TEE98 pKa = 4.13EE99 pKa = 3.68EE100 pKa = 4.31SRR102 pKa = 11.84PSASEE107 pKa = 3.81VANATQRR114 pKa = 11.84VDD116 pKa = 3.42DD117 pKa = 3.9ATVAIRR123 pKa = 11.84SQIQLLLNEE132 pKa = 4.95LSNGHH137 pKa = 5.86GLMNRR142 pKa = 11.84AEE144 pKa = 4.44FEE146 pKa = 4.33VLLPWATAPATT157 pKa = 3.61
MM1 pKa = 7.52VYY3 pKa = 10.67NITSSNQYY11 pKa = 10.63QYY13 pKa = 10.8FAAMWAEE20 pKa = 4.06PTAMLNQCVSALSQSYY36 pKa = 7.22QTQAARR42 pKa = 11.84DD43 pKa = 3.67TVRR46 pKa = 11.84QQFSNLLSAIVTPNQRR62 pKa = 11.84FPEE65 pKa = 4.32AGYY68 pKa = 10.16RR69 pKa = 11.84VYY71 pKa = 10.85INSAVLKK78 pKa = 9.44PLYY81 pKa = 9.69EE82 pKa = 4.29SLMKK86 pKa = 10.92SFDD89 pKa = 3.23TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.26TEE98 pKa = 4.13EE99 pKa = 3.68EE100 pKa = 4.31SRR102 pKa = 11.84PSASEE107 pKa = 3.81VANATQRR114 pKa = 11.84VDD116 pKa = 3.42DD117 pKa = 3.9ATVAIRR123 pKa = 11.84SQIQLLLNEE132 pKa = 4.95LSNGHH137 pKa = 5.86GLMNRR142 pKa = 11.84AEE144 pKa = 4.44FEE146 pKa = 4.33VLLPWATAPATT157 pKa = 3.61
Molecular weight: 17.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q66222|CAPSD_YOMV Capsid protein OS=Youcai mosaic virus OX=228578 GN=CP PE=2 SV=3
MM1 pKa = 7.94SYY3 pKa = 10.28EE4 pKa = 4.08PKK6 pKa = 10.69VSDD9 pKa = 4.73FLALTKK15 pKa = 10.38KK16 pKa = 10.85EE17 pKa = 4.14EE18 pKa = 4.2ILPKK22 pKa = 10.55ALTRR26 pKa = 11.84LKK28 pKa = 9.72TVSISTKK35 pKa = 10.4DD36 pKa = 3.46VISVKK41 pKa = 10.08EE42 pKa = 4.16SEE44 pKa = 4.5SLCDD48 pKa = 3.89IDD50 pKa = 6.04LLVNVPLDD58 pKa = 3.17KK59 pKa = 10.78YY60 pKa = 10.67RR61 pKa = 11.84YY62 pKa = 8.15VGVLGVVFTGEE73 pKa = 3.77WLVPDD78 pKa = 4.61FVKK81 pKa = 10.99GGVTVSVIDD90 pKa = 4.22KK91 pKa = 10.23RR92 pKa = 11.84LEE94 pKa = 3.82NSKK97 pKa = 10.48EE98 pKa = 4.22CIIGTYY104 pKa = 9.64RR105 pKa = 11.84AAAKK109 pKa = 10.17DD110 pKa = 3.26RR111 pKa = 11.84RR112 pKa = 11.84FQFKK116 pKa = 10.22LVPNYY121 pKa = 9.44FVSVADD127 pKa = 4.32AKK129 pKa = 10.32RR130 pKa = 11.84KK131 pKa = 7.58PWQVHH136 pKa = 4.64VRR138 pKa = 11.84IQNLKK143 pKa = 9.94IEE145 pKa = 5.03AGWQPLALEE154 pKa = 4.52VVSVAMVTNNVVVKK168 pKa = 9.49GLRR171 pKa = 11.84EE172 pKa = 3.73KK173 pKa = 10.91VIAVNDD179 pKa = 4.06PNVEE183 pKa = 4.0GFEE186 pKa = 4.26GVVDD190 pKa = 4.18DD191 pKa = 5.83FVDD194 pKa = 3.91SVAAFKK200 pKa = 10.9AIDD203 pKa = 3.49SFRR206 pKa = 11.84KK207 pKa = 8.15KK208 pKa = 10.03KK209 pKa = 10.47KK210 pKa = 10.04RR211 pKa = 11.84IGGRR215 pKa = 11.84DD216 pKa = 3.46VNSNKK221 pKa = 9.53YY222 pKa = 8.94RR223 pKa = 11.84YY224 pKa = 9.6RR225 pKa = 11.84PEE227 pKa = 5.01RR228 pKa = 11.84YY229 pKa = 9.44AGPDD233 pKa = 2.87SLQYY237 pKa = 10.63KK238 pKa = 9.31EE239 pKa = 4.9EE240 pKa = 4.37NGLQHH245 pKa = 6.97HH246 pKa = 6.38EE247 pKa = 4.32LEE249 pKa = 4.52SVPVFRR255 pKa = 11.84SDD257 pKa = 2.84VGRR260 pKa = 11.84AHH262 pKa = 7.36SDD264 pKa = 2.78AA265 pKa = 5.51
MM1 pKa = 7.94SYY3 pKa = 10.28EE4 pKa = 4.08PKK6 pKa = 10.69VSDD9 pKa = 4.73FLALTKK15 pKa = 10.38KK16 pKa = 10.85EE17 pKa = 4.14EE18 pKa = 4.2ILPKK22 pKa = 10.55ALTRR26 pKa = 11.84LKK28 pKa = 9.72TVSISTKK35 pKa = 10.4DD36 pKa = 3.46VISVKK41 pKa = 10.08EE42 pKa = 4.16SEE44 pKa = 4.5SLCDD48 pKa = 3.89IDD50 pKa = 6.04LLVNVPLDD58 pKa = 3.17KK59 pKa = 10.78YY60 pKa = 10.67RR61 pKa = 11.84YY62 pKa = 8.15VGVLGVVFTGEE73 pKa = 3.77WLVPDD78 pKa = 4.61FVKK81 pKa = 10.99GGVTVSVIDD90 pKa = 4.22KK91 pKa = 10.23RR92 pKa = 11.84LEE94 pKa = 3.82NSKK97 pKa = 10.48EE98 pKa = 4.22CIIGTYY104 pKa = 9.64RR105 pKa = 11.84AAAKK109 pKa = 10.17DD110 pKa = 3.26RR111 pKa = 11.84RR112 pKa = 11.84FQFKK116 pKa = 10.22LVPNYY121 pKa = 9.44FVSVADD127 pKa = 4.32AKK129 pKa = 10.32RR130 pKa = 11.84KK131 pKa = 7.58PWQVHH136 pKa = 4.64VRR138 pKa = 11.84IQNLKK143 pKa = 9.94IEE145 pKa = 5.03AGWQPLALEE154 pKa = 4.52VVSVAMVTNNVVVKK168 pKa = 9.49GLRR171 pKa = 11.84EE172 pKa = 3.73KK173 pKa = 10.91VIAVNDD179 pKa = 4.06PNVEE183 pKa = 4.0GFEE186 pKa = 4.26GVVDD190 pKa = 4.18DD191 pKa = 5.83FVDD194 pKa = 3.91SVAAFKK200 pKa = 10.9AIDD203 pKa = 3.49SFRR206 pKa = 11.84KK207 pKa = 8.15KK208 pKa = 10.03KK209 pKa = 10.47KK210 pKa = 10.04RR211 pKa = 11.84IGGRR215 pKa = 11.84DD216 pKa = 3.46VNSNKK221 pKa = 9.53YY222 pKa = 8.94RR223 pKa = 11.84YY224 pKa = 9.6RR225 pKa = 11.84PEE227 pKa = 5.01RR228 pKa = 11.84YY229 pKa = 9.44AGPDD233 pKa = 2.87SLQYY237 pKa = 10.63KK238 pKa = 9.31EE239 pKa = 4.9EE240 pKa = 4.37NGLQHH245 pKa = 6.97HH246 pKa = 6.38EE247 pKa = 4.32LEE249 pKa = 4.52SVPVFRR255 pKa = 11.84SDD257 pKa = 2.84VGRR260 pKa = 11.84AHH262 pKa = 7.36SDD264 pKa = 2.78AA265 pKa = 5.51
Molecular weight: 29.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2019 |
157 |
1597 |
673.0 |
76.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.33 ± 0.884 | 1.981 ± 0.609 |
6.241 ± 0.697 | 6.439 ± 0.207 |
5.052 ± 0.605 | 4.408 ± 0.482 |
2.13 ± 0.456 | 4.755 ± 0.121 |
7.083 ± 1.07 | 9.411 ± 0.692 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.179 ± 0.422 | 4.26 ± 0.557 |
3.814 ± 0.216 | 3.269 ± 0.801 |
5.25 ± 0.552 | 6.786 ± 0.58 |
5.745 ± 0.83 | 8.866 ± 1.668 |
1.189 ± 0.019 | 3.764 ± 0.136 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
