
Hubei sobemo-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.39
Get precalculated fractions of proteins
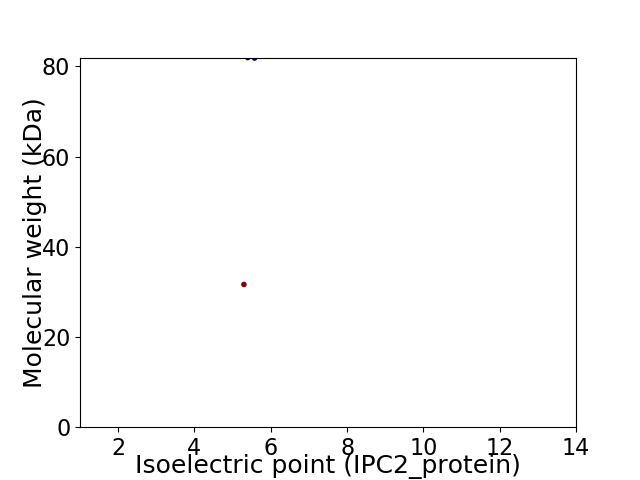
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEP6|A0A1L3KEP6_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 5 OX=1923238 PE=4 SV=1
MM1 pKa = 7.48VDD3 pKa = 3.37RR4 pKa = 11.84VLFGPWSAADD14 pKa = 3.25TATAMEE20 pKa = 4.39RR21 pKa = 11.84AGKK24 pKa = 10.01SGWSLIPNGFYY35 pKa = 10.96DD36 pKa = 4.37LLAYY40 pKa = 10.45FPDD43 pKa = 4.73DD44 pKa = 4.56VLATDD49 pKa = 4.97CSAFDD54 pKa = 3.34WTYY57 pKa = 9.32PAWVIDD63 pKa = 4.17PIIDD67 pKa = 3.59VKK69 pKa = 10.81LEE71 pKa = 4.11QMGSLPDD78 pKa = 4.27GYY80 pKa = 10.11ATAVRR85 pKa = 11.84TRR87 pKa = 11.84ITEE90 pKa = 4.01VLGVNCTLRR99 pKa = 11.84LPDD102 pKa = 3.82GQRR105 pKa = 11.84LRR107 pKa = 11.84QLVPGIMKK115 pKa = 10.37SGWFLTINLNSDD127 pKa = 3.6AQDD130 pKa = 3.26MLTTLAYY137 pKa = 9.57RR138 pKa = 11.84RR139 pKa = 11.84AYY141 pKa = 10.32GSEE144 pKa = 4.33CPLLWAMGDD153 pKa = 3.71DD154 pKa = 4.38VLMRR158 pKa = 11.84WPQGQDD164 pKa = 2.79SAPLVQQLRR173 pKa = 11.84RR174 pKa = 11.84AGILSKK180 pKa = 10.45FATPSRR186 pKa = 11.84EE187 pKa = 3.64FSGFAVKK194 pKa = 9.37RR195 pKa = 11.84TGSDD199 pKa = 2.59IAVNPLYY206 pKa = 10.04PNKK209 pKa = 10.14HH210 pKa = 6.22KK211 pKa = 10.98YY212 pKa = 10.05LLAHH216 pKa = 6.54TSATDD221 pKa = 3.57LEE223 pKa = 4.42EE224 pKa = 4.64VITSFGMIYY233 pKa = 10.75ALAEE237 pKa = 4.39PDD239 pKa = 3.32VKK241 pKa = 11.0AWLEE245 pKa = 3.78PLLRR249 pKa = 11.84QYY251 pKa = 11.24SRR253 pKa = 11.84WPQSSFRR260 pKa = 11.84AWAHH264 pKa = 5.73GLLSHH269 pKa = 7.24APLLQTGDD277 pKa = 3.51AAGSFGLEE285 pKa = 3.84
MM1 pKa = 7.48VDD3 pKa = 3.37RR4 pKa = 11.84VLFGPWSAADD14 pKa = 3.25TATAMEE20 pKa = 4.39RR21 pKa = 11.84AGKK24 pKa = 10.01SGWSLIPNGFYY35 pKa = 10.96DD36 pKa = 4.37LLAYY40 pKa = 10.45FPDD43 pKa = 4.73DD44 pKa = 4.56VLATDD49 pKa = 4.97CSAFDD54 pKa = 3.34WTYY57 pKa = 9.32PAWVIDD63 pKa = 4.17PIIDD67 pKa = 3.59VKK69 pKa = 10.81LEE71 pKa = 4.11QMGSLPDD78 pKa = 4.27GYY80 pKa = 10.11ATAVRR85 pKa = 11.84TRR87 pKa = 11.84ITEE90 pKa = 4.01VLGVNCTLRR99 pKa = 11.84LPDD102 pKa = 3.82GQRR105 pKa = 11.84LRR107 pKa = 11.84QLVPGIMKK115 pKa = 10.37SGWFLTINLNSDD127 pKa = 3.6AQDD130 pKa = 3.26MLTTLAYY137 pKa = 9.57RR138 pKa = 11.84RR139 pKa = 11.84AYY141 pKa = 10.32GSEE144 pKa = 4.33CPLLWAMGDD153 pKa = 3.71DD154 pKa = 4.38VLMRR158 pKa = 11.84WPQGQDD164 pKa = 2.79SAPLVQQLRR173 pKa = 11.84RR174 pKa = 11.84AGILSKK180 pKa = 10.45FATPSRR186 pKa = 11.84EE187 pKa = 3.64FSGFAVKK194 pKa = 9.37RR195 pKa = 11.84TGSDD199 pKa = 2.59IAVNPLYY206 pKa = 10.04PNKK209 pKa = 10.14HH210 pKa = 6.22KK211 pKa = 10.98YY212 pKa = 10.05LLAHH216 pKa = 6.54TSATDD221 pKa = 3.57LEE223 pKa = 4.42EE224 pKa = 4.64VITSFGMIYY233 pKa = 10.75ALAEE237 pKa = 4.39PDD239 pKa = 3.32VKK241 pKa = 11.0AWLEE245 pKa = 3.78PLLRR249 pKa = 11.84QYY251 pKa = 11.24SRR253 pKa = 11.84WPQSSFRR260 pKa = 11.84AWAHH264 pKa = 5.73GLLSHH269 pKa = 7.24APLLQTGDD277 pKa = 3.51AAGSFGLEE285 pKa = 3.84
Molecular weight: 31.64 kDa
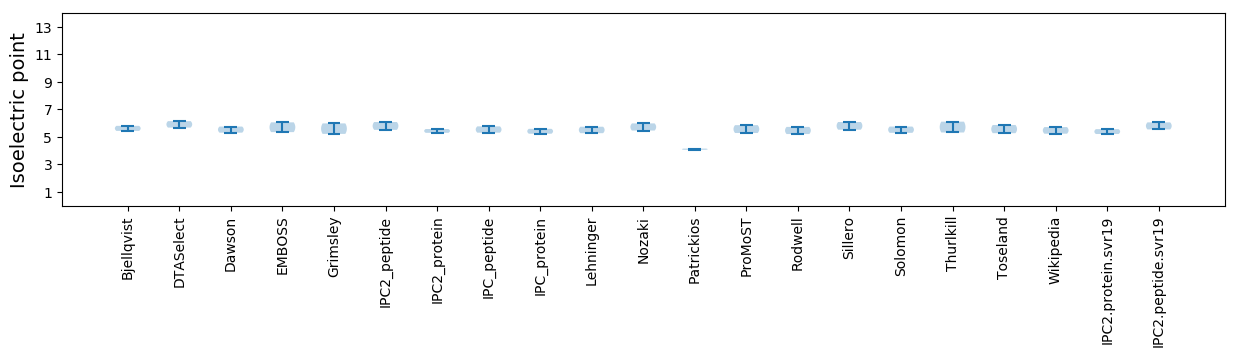
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEP6|A0A1L3KEP6_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 5 OX=1923238 PE=4 SV=1
MM1 pKa = 7.28TPVEE5 pKa = 4.09MFKK8 pKa = 10.8FVIKK12 pKa = 9.9TISDD16 pKa = 3.86FLKK19 pKa = 10.55LLYY22 pKa = 8.98FTAVSMAVLHH32 pKa = 6.04LLIAFSSYY40 pKa = 10.6QFLTSYY46 pKa = 8.99WLCEE50 pKa = 3.94PKK52 pKa = 10.32NRR54 pKa = 11.84SYY56 pKa = 11.52YY57 pKa = 10.61EE58 pKa = 3.91PLNCLGMLTYY68 pKa = 10.05WGFGEE73 pKa = 4.32VLLFATFIAYY83 pKa = 9.84CLLAWFNSPPRR94 pKa = 11.84LTFSPEE100 pKa = 3.39DD101 pKa = 3.53AEE103 pKa = 4.16RR104 pKa = 11.84LEE106 pKa = 4.0RR107 pKa = 11.84DD108 pKa = 3.11RR109 pKa = 11.84ARR111 pKa = 11.84RR112 pKa = 11.84GAYY115 pKa = 9.35GSNEE119 pKa = 3.6ALVRR123 pKa = 11.84EE124 pKa = 4.49FMAWAGTPEE133 pKa = 4.27SYY135 pKa = 10.59KK136 pKa = 10.49EE137 pKa = 3.88DD138 pKa = 3.51SPYY141 pKa = 11.29VSMTPPSWQFSVWRR155 pKa = 11.84AQTEE159 pKa = 4.18AKK161 pKa = 8.29KK162 pKa = 8.9TLIGHH167 pKa = 6.02GCRR170 pKa = 11.84INGHH174 pKa = 6.05LVINYY179 pKa = 8.41HH180 pKa = 5.65VLNTAPLDD188 pKa = 3.72QLYY191 pKa = 10.46LAIIRR196 pKa = 11.84PGKK199 pKa = 7.88EE200 pKa = 3.53TVVQSLATFQFSEE213 pKa = 4.8LLPDD217 pKa = 3.53ICVCPIAQLKK227 pKa = 9.69GIQLTGLKK235 pKa = 8.85EE236 pKa = 4.06AKK238 pKa = 9.53VKK240 pKa = 10.28HH241 pKa = 4.91VQGFQPAYY249 pKa = 9.49IATDD253 pKa = 3.9FPDD256 pKa = 3.93NNASTAAVINHH267 pKa = 6.19PEE269 pKa = 3.66AWGMLQYY276 pKa = 10.68KK277 pKa = 10.42GSTRR281 pKa = 11.84PGFSGATYY289 pKa = 10.36VHH291 pKa = 6.83GSSLFGIHH299 pKa = 5.97CHH301 pKa = 6.75GGIQNVGYY309 pKa = 8.89SASYY313 pKa = 10.17IALKK317 pKa = 10.55LKK319 pKa = 10.74NNEE322 pKa = 3.78SSDD325 pKa = 4.07YY326 pKa = 10.74YY327 pKa = 11.35ALQAMLRR334 pKa = 11.84SSRR337 pKa = 11.84DD338 pKa = 2.59RR339 pKa = 11.84DD340 pKa = 3.73YY341 pKa = 11.46QSQRR345 pKa = 11.84VNPDD349 pKa = 2.49EE350 pKa = 4.47FEE352 pKa = 4.04IRR354 pKa = 11.84FQGRR358 pKa = 11.84YY359 pKa = 9.31FIIEE363 pKa = 3.92TEE365 pKa = 4.44EE366 pKa = 3.99YY367 pKa = 10.63QEE369 pKa = 5.91LEE371 pKa = 4.29DD372 pKa = 4.4NYY374 pKa = 11.44GDD376 pKa = 4.25GSDD379 pKa = 3.4FHH381 pKa = 6.96PRR383 pKa = 11.84SKK385 pKa = 10.64KK386 pKa = 9.44KK387 pKa = 10.29AWRR390 pKa = 11.84NRR392 pKa = 11.84QDD394 pKa = 3.07WEE396 pKa = 4.25NNTAFDD402 pKa = 4.29PMLLPWEE409 pKa = 4.44NNNVSPSSPSQQPPSTTEE427 pKa = 3.25IATQTEE433 pKa = 4.74MEE435 pKa = 4.46WEE437 pKa = 4.08NAPVFDD443 pKa = 5.45DD444 pKa = 4.44GFRR447 pKa = 11.84IQQLTARR454 pKa = 11.84VDD456 pKa = 3.51ALMAQCDD463 pKa = 3.87SEE465 pKa = 4.95RR466 pKa = 11.84EE467 pKa = 3.7QRR469 pKa = 11.84LALRR473 pKa = 11.84QQCEE477 pKa = 4.24RR478 pKa = 11.84EE479 pKa = 4.11WANTTMWIEE488 pKa = 3.99ALNDD492 pKa = 3.58KK493 pKa = 10.46IMRR496 pKa = 11.84LWDD499 pKa = 3.66RR500 pKa = 11.84EE501 pKa = 4.16LGLEE505 pKa = 5.07SEE507 pKa = 5.37DD508 pKa = 3.67EE509 pKa = 4.26DD510 pKa = 5.17DD511 pKa = 4.7PRR513 pKa = 11.84EE514 pKa = 4.09LFPGVVFEE522 pKa = 4.72NADD525 pKa = 3.64PEE527 pKa = 4.42PTVPEE532 pKa = 3.87IEE534 pKa = 3.93ITEE537 pKa = 4.28VVEE540 pKa = 4.63PEE542 pKa = 3.99APNANSPTQNTMGGATSGMQEE563 pKa = 4.39PTSSHH568 pKa = 6.44PPSPNQAHH576 pKa = 7.27LSTSQMDD583 pKa = 3.84SLISGLQQHH592 pKa = 5.59QQQSARR598 pKa = 11.84ALQSIQSILAQQTALLSRR616 pKa = 11.84LTPEE620 pKa = 3.68NHH622 pKa = 6.3SSQEE626 pKa = 4.02RR627 pKa = 11.84GSGSGTAPVTRR638 pKa = 11.84RR639 pKa = 11.84GNRR642 pKa = 11.84WSRR645 pKa = 11.84RR646 pKa = 11.84TRR648 pKa = 11.84RR649 pKa = 11.84PGSRR653 pKa = 11.84QPGASTGPEE662 pKa = 3.96QQASAPSRR670 pKa = 11.84VTQPSRR676 pKa = 11.84TRR678 pKa = 11.84LDD680 pKa = 3.45GTASASPTVRR690 pKa = 11.84TGRR693 pKa = 11.84SSSNSSQPALTPSQRR708 pKa = 11.84AAAAALRR715 pKa = 11.84RR716 pKa = 11.84NPRR719 pKa = 11.84VQPAQQTSSSSS730 pKa = 3.11
MM1 pKa = 7.28TPVEE5 pKa = 4.09MFKK8 pKa = 10.8FVIKK12 pKa = 9.9TISDD16 pKa = 3.86FLKK19 pKa = 10.55LLYY22 pKa = 8.98FTAVSMAVLHH32 pKa = 6.04LLIAFSSYY40 pKa = 10.6QFLTSYY46 pKa = 8.99WLCEE50 pKa = 3.94PKK52 pKa = 10.32NRR54 pKa = 11.84SYY56 pKa = 11.52YY57 pKa = 10.61EE58 pKa = 3.91PLNCLGMLTYY68 pKa = 10.05WGFGEE73 pKa = 4.32VLLFATFIAYY83 pKa = 9.84CLLAWFNSPPRR94 pKa = 11.84LTFSPEE100 pKa = 3.39DD101 pKa = 3.53AEE103 pKa = 4.16RR104 pKa = 11.84LEE106 pKa = 4.0RR107 pKa = 11.84DD108 pKa = 3.11RR109 pKa = 11.84ARR111 pKa = 11.84RR112 pKa = 11.84GAYY115 pKa = 9.35GSNEE119 pKa = 3.6ALVRR123 pKa = 11.84EE124 pKa = 4.49FMAWAGTPEE133 pKa = 4.27SYY135 pKa = 10.59KK136 pKa = 10.49EE137 pKa = 3.88DD138 pKa = 3.51SPYY141 pKa = 11.29VSMTPPSWQFSVWRR155 pKa = 11.84AQTEE159 pKa = 4.18AKK161 pKa = 8.29KK162 pKa = 8.9TLIGHH167 pKa = 6.02GCRR170 pKa = 11.84INGHH174 pKa = 6.05LVINYY179 pKa = 8.41HH180 pKa = 5.65VLNTAPLDD188 pKa = 3.72QLYY191 pKa = 10.46LAIIRR196 pKa = 11.84PGKK199 pKa = 7.88EE200 pKa = 3.53TVVQSLATFQFSEE213 pKa = 4.8LLPDD217 pKa = 3.53ICVCPIAQLKK227 pKa = 9.69GIQLTGLKK235 pKa = 8.85EE236 pKa = 4.06AKK238 pKa = 9.53VKK240 pKa = 10.28HH241 pKa = 4.91VQGFQPAYY249 pKa = 9.49IATDD253 pKa = 3.9FPDD256 pKa = 3.93NNASTAAVINHH267 pKa = 6.19PEE269 pKa = 3.66AWGMLQYY276 pKa = 10.68KK277 pKa = 10.42GSTRR281 pKa = 11.84PGFSGATYY289 pKa = 10.36VHH291 pKa = 6.83GSSLFGIHH299 pKa = 5.97CHH301 pKa = 6.75GGIQNVGYY309 pKa = 8.89SASYY313 pKa = 10.17IALKK317 pKa = 10.55LKK319 pKa = 10.74NNEE322 pKa = 3.78SSDD325 pKa = 4.07YY326 pKa = 10.74YY327 pKa = 11.35ALQAMLRR334 pKa = 11.84SSRR337 pKa = 11.84DD338 pKa = 2.59RR339 pKa = 11.84DD340 pKa = 3.73YY341 pKa = 11.46QSQRR345 pKa = 11.84VNPDD349 pKa = 2.49EE350 pKa = 4.47FEE352 pKa = 4.04IRR354 pKa = 11.84FQGRR358 pKa = 11.84YY359 pKa = 9.31FIIEE363 pKa = 3.92TEE365 pKa = 4.44EE366 pKa = 3.99YY367 pKa = 10.63QEE369 pKa = 5.91LEE371 pKa = 4.29DD372 pKa = 4.4NYY374 pKa = 11.44GDD376 pKa = 4.25GSDD379 pKa = 3.4FHH381 pKa = 6.96PRR383 pKa = 11.84SKK385 pKa = 10.64KK386 pKa = 9.44KK387 pKa = 10.29AWRR390 pKa = 11.84NRR392 pKa = 11.84QDD394 pKa = 3.07WEE396 pKa = 4.25NNTAFDD402 pKa = 4.29PMLLPWEE409 pKa = 4.44NNNVSPSSPSQQPPSTTEE427 pKa = 3.25IATQTEE433 pKa = 4.74MEE435 pKa = 4.46WEE437 pKa = 4.08NAPVFDD443 pKa = 5.45DD444 pKa = 4.44GFRR447 pKa = 11.84IQQLTARR454 pKa = 11.84VDD456 pKa = 3.51ALMAQCDD463 pKa = 3.87SEE465 pKa = 4.95RR466 pKa = 11.84EE467 pKa = 3.7QRR469 pKa = 11.84LALRR473 pKa = 11.84QQCEE477 pKa = 4.24RR478 pKa = 11.84EE479 pKa = 4.11WANTTMWIEE488 pKa = 3.99ALNDD492 pKa = 3.58KK493 pKa = 10.46IMRR496 pKa = 11.84LWDD499 pKa = 3.66RR500 pKa = 11.84EE501 pKa = 4.16LGLEE505 pKa = 5.07SEE507 pKa = 5.37DD508 pKa = 3.67EE509 pKa = 4.26DD510 pKa = 5.17DD511 pKa = 4.7PRR513 pKa = 11.84EE514 pKa = 4.09LFPGVVFEE522 pKa = 4.72NADD525 pKa = 3.64PEE527 pKa = 4.42PTVPEE532 pKa = 3.87IEE534 pKa = 3.93ITEE537 pKa = 4.28VVEE540 pKa = 4.63PEE542 pKa = 3.99APNANSPTQNTMGGATSGMQEE563 pKa = 4.39PTSSHH568 pKa = 6.44PPSPNQAHH576 pKa = 7.27LSTSQMDD583 pKa = 3.84SLISGLQQHH592 pKa = 5.59QQQSARR598 pKa = 11.84ALQSIQSILAQQTALLSRR616 pKa = 11.84LTPEE620 pKa = 3.68NHH622 pKa = 6.3SSQEE626 pKa = 4.02RR627 pKa = 11.84GSGSGTAPVTRR638 pKa = 11.84RR639 pKa = 11.84GNRR642 pKa = 11.84WSRR645 pKa = 11.84RR646 pKa = 11.84TRR648 pKa = 11.84RR649 pKa = 11.84PGSRR653 pKa = 11.84QPGASTGPEE662 pKa = 3.96QQASAPSRR670 pKa = 11.84VTQPSRR676 pKa = 11.84TRR678 pKa = 11.84LDD680 pKa = 3.45GTASASPTVRR690 pKa = 11.84TGRR693 pKa = 11.84SSSNSSQPALTPSQRR708 pKa = 11.84AAAAALRR715 pKa = 11.84RR716 pKa = 11.84NPRR719 pKa = 11.84VQPAQQTSSSSS730 pKa = 3.11
Molecular weight: 81.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1015 |
285 |
730 |
507.5 |
56.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.064 ± 0.758 | 1.182 ± 0.067 |
4.828 ± 1.136 | 6.01 ± 1.297 |
3.842 ± 0.009 | 6.108 ± 0.653 |
1.773 ± 0.192 | 3.941 ± 0.042 |
2.66 ± 0.076 | 9.261 ± 1.566 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.229 | 3.842 ± 0.901 |
6.798 ± 0.25 | 6.01 ± 1.115 |
6.601 ± 0.33 | 9.064 ± 1.061 |
6.404 ± 0.228 | 4.532 ± 0.379 |
2.463 ± 0.542 | 3.251 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
