
Smittium megazygosporum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Zoopagomycota; Kickxellomycotina; Harpellomycetes; Harpellales; Legeriomycetaceae; Smittium
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
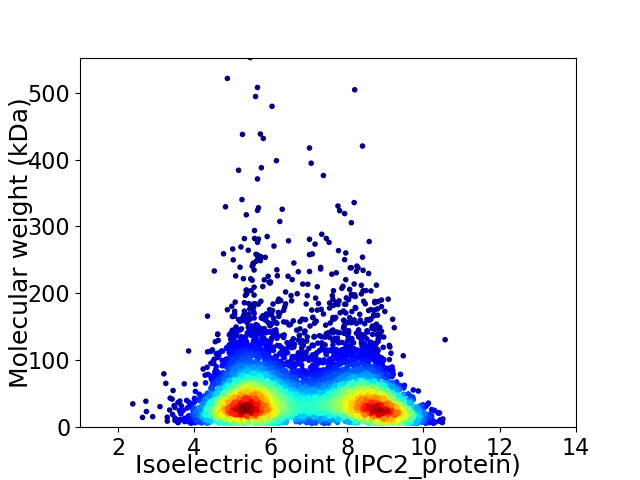
Virtual 2D-PAGE plot for 6835 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2T9ZD40|A0A2T9ZD40_9FUNG Uncharacterized protein OS=Smittium megazygosporum OX=133381 GN=BB560_003038 PE=4 SV=1
MM1 pKa = 7.75LILFLILVLFGAQNISAQNEE21 pKa = 4.04FSCVNGRR28 pKa = 11.84AVYY31 pKa = 9.06TYY33 pKa = 10.44DD34 pKa = 4.34GNITLSGDD42 pKa = 3.62PNGGTDD48 pKa = 4.97AFIKK52 pKa = 10.12DD53 pKa = 3.97LSMLCDD59 pKa = 3.38SPSFQITVTGYY70 pKa = 10.51GGSDD74 pKa = 3.44PLISLHH80 pKa = 5.84NQASSADD87 pKa = 4.53AIYY90 pKa = 9.9MDD92 pKa = 4.04YY93 pKa = 11.07GFEE96 pKa = 3.88SGYY99 pKa = 11.01YY100 pKa = 8.92SVCIKK105 pKa = 10.7QSDD108 pKa = 3.81SMGACTKK115 pKa = 8.58TQSTKK120 pKa = 10.65YY121 pKa = 9.77VNKK124 pKa = 9.19LTTIYY129 pKa = 11.01VNVNGTIIQMGTVTVKK145 pKa = 10.57GVNTKK150 pKa = 8.23ITQTTFPSGYY160 pKa = 8.54NVDD163 pKa = 4.1YY164 pKa = 10.96IRR166 pKa = 11.84FSVWEE171 pKa = 4.03SLFAYY176 pKa = 9.29TNIQLRR182 pKa = 11.84CDD184 pKa = 4.88DD185 pKa = 3.88ICDD188 pKa = 3.49NSTVSNKK195 pKa = 6.62TWKK198 pKa = 9.9PIPTTSSTTPVLTTSAPTTTATPALTTSAPITTATTLTTPTTPVLTTSDD247 pKa = 4.48SITTANPLNTTTTSITTSVIPTSSSAIISLTTTIGIDD284 pKa = 3.41VSISDD289 pKa = 3.64TAMITAEE296 pKa = 4.18TTFDD300 pKa = 3.17VSTISTGDD308 pKa = 3.28TKK310 pKa = 11.6SNTNLNNITTNTYY323 pKa = 10.25TMVDD327 pKa = 3.34TALTSTIYY335 pKa = 7.9TTTDD339 pKa = 2.71TAAGTDD345 pKa = 3.15RR346 pKa = 11.84GIISTGLTTTEE357 pKa = 3.97TDD359 pKa = 3.19SFSASTGISSADD371 pKa = 3.15TGAPGTDD378 pKa = 2.74TSVARR383 pKa = 11.84TTSGIDD389 pKa = 3.06EE390 pKa = 4.79TNTVDD395 pKa = 2.93TSTISTVLVDD405 pKa = 3.36TSTNYY410 pKa = 10.4SYY412 pKa = 11.32SVSSSTGTNIIISTEE427 pKa = 4.04YY428 pKa = 10.2TEE430 pKa = 5.1SSSTTITSGTGAPDD444 pKa = 2.89ITGFTDD450 pKa = 3.15TNTVADD456 pKa = 3.85ITVSASTEE464 pKa = 3.91AKK466 pKa = 9.02STVSSSTQTLITTGSTGSIGTTDD489 pKa = 3.67SFNGPTDD496 pKa = 3.31TRR498 pKa = 11.84TMSTSATEE506 pKa = 4.06STYY509 pKa = 10.45LTDD512 pKa = 5.22IYY514 pKa = 9.69TQSGNTTEE522 pKa = 5.25SEE524 pKa = 4.4TTNLTNIDD532 pKa = 4.11TEE534 pKa = 4.53STSAGATSLIGATEE548 pKa = 4.35SNNSEE553 pKa = 4.15STGKK557 pKa = 10.92SNTTSLTNGNTEE569 pKa = 4.18STNSSMTEE577 pKa = 3.61EE578 pKa = 4.49TIATSSIGTEE588 pKa = 4.01DD589 pKa = 3.38RR590 pKa = 11.84TTGTGATDD598 pKa = 3.67TGATGTLTGATDD610 pKa = 3.74TGATGTLTGATDD622 pKa = 3.74TGATGTLTT630 pKa = 3.45
MM1 pKa = 7.75LILFLILVLFGAQNISAQNEE21 pKa = 4.04FSCVNGRR28 pKa = 11.84AVYY31 pKa = 9.06TYY33 pKa = 10.44DD34 pKa = 4.34GNITLSGDD42 pKa = 3.62PNGGTDD48 pKa = 4.97AFIKK52 pKa = 10.12DD53 pKa = 3.97LSMLCDD59 pKa = 3.38SPSFQITVTGYY70 pKa = 10.51GGSDD74 pKa = 3.44PLISLHH80 pKa = 5.84NQASSADD87 pKa = 4.53AIYY90 pKa = 9.9MDD92 pKa = 4.04YY93 pKa = 11.07GFEE96 pKa = 3.88SGYY99 pKa = 11.01YY100 pKa = 8.92SVCIKK105 pKa = 10.7QSDD108 pKa = 3.81SMGACTKK115 pKa = 8.58TQSTKK120 pKa = 10.65YY121 pKa = 9.77VNKK124 pKa = 9.19LTTIYY129 pKa = 11.01VNVNGTIIQMGTVTVKK145 pKa = 10.57GVNTKK150 pKa = 8.23ITQTTFPSGYY160 pKa = 8.54NVDD163 pKa = 4.1YY164 pKa = 10.96IRR166 pKa = 11.84FSVWEE171 pKa = 4.03SLFAYY176 pKa = 9.29TNIQLRR182 pKa = 11.84CDD184 pKa = 4.88DD185 pKa = 3.88ICDD188 pKa = 3.49NSTVSNKK195 pKa = 6.62TWKK198 pKa = 9.9PIPTTSSTTPVLTTSAPTTTATPALTTSAPITTATTLTTPTTPVLTTSDD247 pKa = 4.48SITTANPLNTTTTSITTSVIPTSSSAIISLTTTIGIDD284 pKa = 3.41VSISDD289 pKa = 3.64TAMITAEE296 pKa = 4.18TTFDD300 pKa = 3.17VSTISTGDD308 pKa = 3.28TKK310 pKa = 11.6SNTNLNNITTNTYY323 pKa = 10.25TMVDD327 pKa = 3.34TALTSTIYY335 pKa = 7.9TTTDD339 pKa = 2.71TAAGTDD345 pKa = 3.15RR346 pKa = 11.84GIISTGLTTTEE357 pKa = 3.97TDD359 pKa = 3.19SFSASTGISSADD371 pKa = 3.15TGAPGTDD378 pKa = 2.74TSVARR383 pKa = 11.84TTSGIDD389 pKa = 3.06EE390 pKa = 4.79TNTVDD395 pKa = 2.93TSTISTVLVDD405 pKa = 3.36TSTNYY410 pKa = 10.4SYY412 pKa = 11.32SVSSSTGTNIIISTEE427 pKa = 4.04YY428 pKa = 10.2TEE430 pKa = 5.1SSSTTITSGTGAPDD444 pKa = 2.89ITGFTDD450 pKa = 3.15TNTVADD456 pKa = 3.85ITVSASTEE464 pKa = 3.91AKK466 pKa = 9.02STVSSSTQTLITTGSTGSIGTTDD489 pKa = 3.67SFNGPTDD496 pKa = 3.31TRR498 pKa = 11.84TMSTSATEE506 pKa = 4.06STYY509 pKa = 10.45LTDD512 pKa = 5.22IYY514 pKa = 9.69TQSGNTTEE522 pKa = 5.25SEE524 pKa = 4.4TTNLTNIDD532 pKa = 4.11TEE534 pKa = 4.53STSAGATSLIGATEE548 pKa = 4.35SNNSEE553 pKa = 4.15STGKK557 pKa = 10.92SNTTSLTNGNTEE569 pKa = 4.18STNSSMTEE577 pKa = 3.61EE578 pKa = 4.49TIATSSIGTEE588 pKa = 4.01DD589 pKa = 3.38RR590 pKa = 11.84TTGTGATDD598 pKa = 3.67TGATGTLTGATDD610 pKa = 3.74TGATGTLTGATDD622 pKa = 3.74TGATGTLTT630 pKa = 3.45
Molecular weight: 64.77 kDa
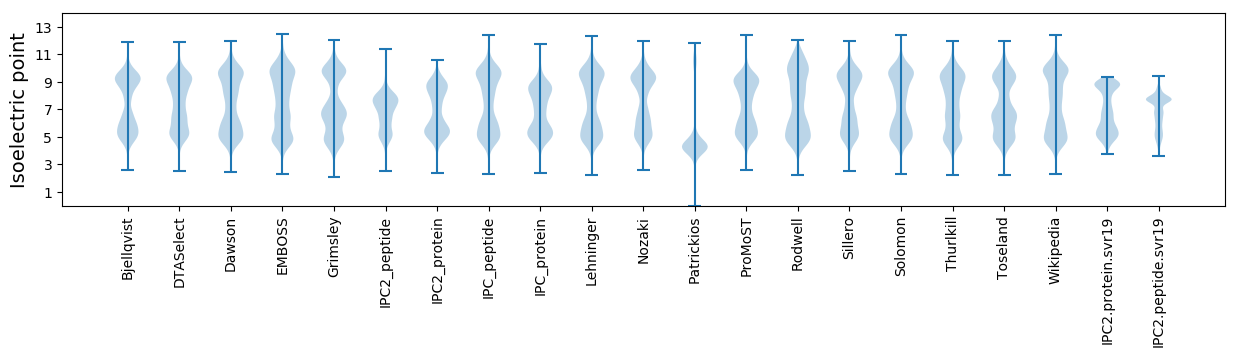
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T9ZAZ1|A0A2T9ZAZ1_9FUNG ABC transporter domain-containing protein OS=Smittium megazygosporum OX=133381 GN=BB560_003791 PE=4 SV=1
MM1 pKa = 6.44TVKK4 pKa = 10.26RR5 pKa = 11.84RR6 pKa = 11.84NHH8 pKa = 4.84GRR10 pKa = 11.84NKK12 pKa = 9.9KK13 pKa = 10.13GRR15 pKa = 11.84GKK17 pKa = 9.21VTFIRR22 pKa = 11.84CDD24 pKa = 3.14NCSRR28 pKa = 11.84CCPKK32 pKa = 10.77DD33 pKa = 3.45KK34 pKa = 10.56AIKK37 pKa = 10.36RR38 pKa = 11.84FTIRR42 pKa = 11.84NMVEE46 pKa = 3.45AAGQRR51 pKa = 11.84DD52 pKa = 3.55MADD55 pKa = 2.72ASVYY59 pKa = 7.35EE60 pKa = 4.57TYY62 pKa = 11.02VLPKK66 pKa = 10.24LYY68 pKa = 10.88LKK70 pKa = 9.37MQYY73 pKa = 10.28CVACAIHH80 pKa = 5.39NHH82 pKa = 4.57IVRR85 pKa = 11.84VRR87 pKa = 11.84STEE90 pKa = 3.48GRR92 pKa = 11.84RR93 pKa = 11.84IRR95 pKa = 11.84TPPPRR100 pKa = 11.84VRR102 pKa = 11.84YY103 pKa = 9.99NKK105 pKa = 9.9DD106 pKa = 2.82GKK108 pKa = 10.61KK109 pKa = 10.0INPAVAAQSAA119 pKa = 3.62
MM1 pKa = 6.44TVKK4 pKa = 10.26RR5 pKa = 11.84RR6 pKa = 11.84NHH8 pKa = 4.84GRR10 pKa = 11.84NKK12 pKa = 9.9KK13 pKa = 10.13GRR15 pKa = 11.84GKK17 pKa = 9.21VTFIRR22 pKa = 11.84CDD24 pKa = 3.14NCSRR28 pKa = 11.84CCPKK32 pKa = 10.77DD33 pKa = 3.45KK34 pKa = 10.56AIKK37 pKa = 10.36RR38 pKa = 11.84FTIRR42 pKa = 11.84NMVEE46 pKa = 3.45AAGQRR51 pKa = 11.84DD52 pKa = 3.55MADD55 pKa = 2.72ASVYY59 pKa = 7.35EE60 pKa = 4.57TYY62 pKa = 11.02VLPKK66 pKa = 10.24LYY68 pKa = 10.88LKK70 pKa = 9.37MQYY73 pKa = 10.28CVACAIHH80 pKa = 5.39NHH82 pKa = 4.57IVRR85 pKa = 11.84VRR87 pKa = 11.84STEE90 pKa = 3.48GRR92 pKa = 11.84RR93 pKa = 11.84IRR95 pKa = 11.84TPPPRR100 pKa = 11.84VRR102 pKa = 11.84YY103 pKa = 9.99NKK105 pKa = 9.9DD106 pKa = 2.82GKK108 pKa = 10.61KK109 pKa = 10.0INPAVAAQSAA119 pKa = 3.62
Molecular weight: 13.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3127516 |
49 |
4877 |
457.6 |
51.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.222 ± 0.028 | 1.21 ± 0.012 |
5.423 ± 0.019 | 6.031 ± 0.027 |
4.753 ± 0.022 | 4.858 ± 0.028 |
1.868 ± 0.013 | 6.841 ± 0.021 |
7.515 ± 0.029 | 9.311 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.823 ± 0.011 | 6.623 ± 0.027 |
4.646 ± 0.028 | 3.828 ± 0.02 |
3.984 ± 0.022 | 10.888 ± 0.047 |
5.552 ± 0.028 | 5.317 ± 0.023 |
0.819 ± 0.009 | 3.488 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
