
Phocoena phocoena papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Omikronpapillomavirus; Omikronpapillomavirus 1
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
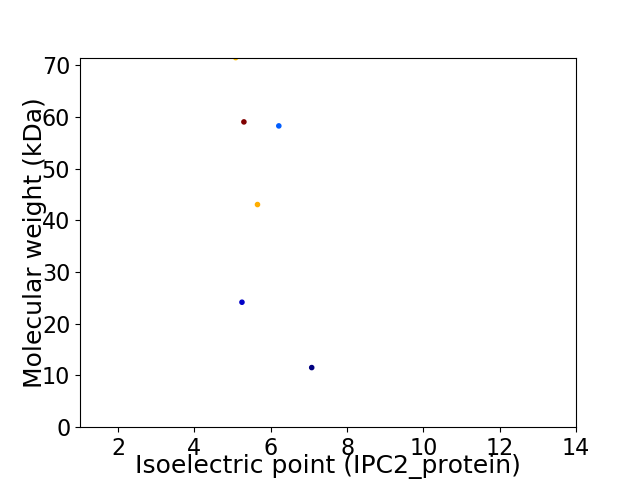
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2VIQ8|F2VIQ8_PSPV Regulatory protein E2 OS=Phocoena phocoena papillomavirus 1 OX=706525 GN=E2 PE=3 SV=1
MM1 pKa = 8.2DD2 pKa = 4.19NTQGTDD8 pKa = 3.38PLEE11 pKa = 4.91GGSGMDD17 pKa = 3.6GGWVLLEE24 pKa = 4.19ATDD27 pKa = 3.56VDD29 pKa = 4.62GAGGDD34 pKa = 3.75DD35 pKa = 6.2DD36 pKa = 5.17EE37 pKa = 7.61DD38 pKa = 4.07EE39 pKa = 5.39DD40 pKa = 4.3EE41 pKa = 6.29DD42 pKa = 4.19YY43 pKa = 11.91GEE45 pKa = 5.4DD46 pKa = 3.51FTDD49 pKa = 6.7FIDD52 pKa = 4.38DD53 pKa = 3.79CAKK56 pKa = 10.03EE57 pKa = 4.25FSEE60 pKa = 4.49EE61 pKa = 4.02THH63 pKa = 6.24TSLFSQQQIEE73 pKa = 4.12EE74 pKa = 4.31DD75 pKa = 3.7DD76 pKa = 4.05RR77 pKa = 11.84AVQALKK83 pKa = 10.59RR84 pKa = 11.84KK85 pKa = 9.09FLEE88 pKa = 4.35SPKK91 pKa = 10.78SKK93 pKa = 10.49VDD95 pKa = 3.6SEE97 pKa = 4.24LSPRR101 pKa = 11.84LAAISLVQRR110 pKa = 11.84TGKK113 pKa = 10.06AKK115 pKa = 10.03KK116 pKa = 9.77RR117 pKa = 11.84LYY119 pKa = 10.32RR120 pKa = 11.84DD121 pKa = 3.21QTQDD125 pKa = 3.05SGHH128 pKa = 6.24GNSEE132 pKa = 3.81EE133 pKa = 4.16SGATEE138 pKa = 4.03VVVSTQVQKK147 pKa = 11.41GGLQRR152 pKa = 11.84VGQTPVVVRR161 pKa = 11.84ASTQQAAEE169 pKa = 4.22SAGSQEE175 pKa = 5.17DD176 pKa = 3.72YY177 pKa = 10.4TGRR180 pKa = 11.84VTQLLRR186 pKa = 11.84SAQPRR191 pKa = 11.84CALLGIFKK199 pKa = 10.44EE200 pKa = 4.33LFACSFMDD208 pKa = 3.73LTRR211 pKa = 11.84CFKK214 pKa = 10.76SDD216 pKa = 3.25STAGEE221 pKa = 4.01DD222 pKa = 3.42WVCFVGGVPCSLADD236 pKa = 3.75GVVDD240 pKa = 5.41LMAPHH245 pKa = 6.54VLYY248 pKa = 11.1SHH250 pKa = 6.82ITSSTCSLGIAVLMLVRR267 pKa = 11.84WKK269 pKa = 8.92TAKK272 pKa = 9.32TRR274 pKa = 11.84ATVKK278 pKa = 10.56KK279 pKa = 10.59LLGSLLAVEE288 pKa = 4.86SGQMLLEE295 pKa = 4.4PPRR298 pKa = 11.84LRR300 pKa = 11.84NPAAAMFWYY309 pKa = 9.49KK310 pKa = 10.81KK311 pKa = 10.5AMCNSTVVTGEE322 pKa = 3.93TPQWILRR329 pKa = 11.84EE330 pKa = 3.98VSIQDD335 pKa = 3.49QVGEE339 pKa = 4.0QCLFSLSDD347 pKa = 3.67MVQWAYY353 pKa = 11.28DD354 pKa = 3.48NGIEE358 pKa = 4.23TEE360 pKa = 4.07SAAAYY365 pKa = 9.04EE366 pKa = 4.25YY367 pKa = 11.58ALLADD372 pKa = 4.03EE373 pKa = 5.67DD374 pKa = 4.38KK375 pKa = 11.52NADD378 pKa = 3.1AFLRR382 pKa = 11.84SNQQAKK388 pKa = 8.18WVGDD392 pKa = 3.59CMRR395 pKa = 11.84MVRR398 pKa = 11.84LYY400 pKa = 10.71RR401 pKa = 11.84RR402 pKa = 11.84AEE404 pKa = 4.03MNKK407 pKa = 7.28MTMGQWIKK415 pKa = 10.45HH416 pKa = 5.29RR417 pKa = 11.84SEE419 pKa = 3.94KK420 pKa = 9.39THH422 pKa = 7.36GEE424 pKa = 4.34GNWKK428 pKa = 10.38EE429 pKa = 3.63IFKK432 pKa = 10.4FLKK435 pKa = 10.04FQGIEE440 pKa = 3.59ITSFLTYY447 pKa = 10.17FRR449 pKa = 11.84LFLKK453 pKa = 10.23GVPKK457 pKa = 10.31KK458 pKa = 10.72NCIAICGPPNTGKK471 pKa = 10.57SLFGMSLIDD480 pKa = 4.62FIDD483 pKa = 3.73GRR485 pKa = 11.84VISHH489 pKa = 6.72CNSNSHH495 pKa = 6.72FWLQPLTEE503 pKa = 4.77CKK505 pKa = 10.13LALLDD510 pKa = 5.16DD511 pKa = 4.53ATPPTWDD518 pKa = 3.48YY519 pKa = 11.59FDD521 pKa = 3.58TYY523 pKa = 11.17LRR525 pKa = 11.84NLADD529 pKa = 4.28GNPVSVDD536 pKa = 3.3TKK538 pKa = 10.81HH539 pKa = 6.94RR540 pKa = 11.84APTQMKK546 pKa = 9.94CPPLLITTNTDD557 pKa = 2.23ISQGDD562 pKa = 3.07RR563 pKa = 11.84WTYY566 pKa = 9.23LKK568 pKa = 10.91SRR570 pKa = 11.84IKK572 pKa = 10.89VFTFPEE578 pKa = 4.07QMPLTEE584 pKa = 4.44MGDD587 pKa = 3.42PAFALTRR594 pKa = 11.84QNWKK598 pKa = 10.67SFFQRR603 pKa = 11.84CWSSLGLEE611 pKa = 4.51DD612 pKa = 5.6PEE614 pKa = 4.28QEE616 pKa = 4.56AANGDD621 pKa = 3.87PRR623 pKa = 11.84EE624 pKa = 4.0VLQPLRR630 pKa = 11.84CTARR634 pKa = 11.84RR635 pKa = 11.84ADD637 pKa = 3.65GLSS640 pKa = 3.1
MM1 pKa = 8.2DD2 pKa = 4.19NTQGTDD8 pKa = 3.38PLEE11 pKa = 4.91GGSGMDD17 pKa = 3.6GGWVLLEE24 pKa = 4.19ATDD27 pKa = 3.56VDD29 pKa = 4.62GAGGDD34 pKa = 3.75DD35 pKa = 6.2DD36 pKa = 5.17EE37 pKa = 7.61DD38 pKa = 4.07EE39 pKa = 5.39DD40 pKa = 4.3EE41 pKa = 6.29DD42 pKa = 4.19YY43 pKa = 11.91GEE45 pKa = 5.4DD46 pKa = 3.51FTDD49 pKa = 6.7FIDD52 pKa = 4.38DD53 pKa = 3.79CAKK56 pKa = 10.03EE57 pKa = 4.25FSEE60 pKa = 4.49EE61 pKa = 4.02THH63 pKa = 6.24TSLFSQQQIEE73 pKa = 4.12EE74 pKa = 4.31DD75 pKa = 3.7DD76 pKa = 4.05RR77 pKa = 11.84AVQALKK83 pKa = 10.59RR84 pKa = 11.84KK85 pKa = 9.09FLEE88 pKa = 4.35SPKK91 pKa = 10.78SKK93 pKa = 10.49VDD95 pKa = 3.6SEE97 pKa = 4.24LSPRR101 pKa = 11.84LAAISLVQRR110 pKa = 11.84TGKK113 pKa = 10.06AKK115 pKa = 10.03KK116 pKa = 9.77RR117 pKa = 11.84LYY119 pKa = 10.32RR120 pKa = 11.84DD121 pKa = 3.21QTQDD125 pKa = 3.05SGHH128 pKa = 6.24GNSEE132 pKa = 3.81EE133 pKa = 4.16SGATEE138 pKa = 4.03VVVSTQVQKK147 pKa = 11.41GGLQRR152 pKa = 11.84VGQTPVVVRR161 pKa = 11.84ASTQQAAEE169 pKa = 4.22SAGSQEE175 pKa = 5.17DD176 pKa = 3.72YY177 pKa = 10.4TGRR180 pKa = 11.84VTQLLRR186 pKa = 11.84SAQPRR191 pKa = 11.84CALLGIFKK199 pKa = 10.44EE200 pKa = 4.33LFACSFMDD208 pKa = 3.73LTRR211 pKa = 11.84CFKK214 pKa = 10.76SDD216 pKa = 3.25STAGEE221 pKa = 4.01DD222 pKa = 3.42WVCFVGGVPCSLADD236 pKa = 3.75GVVDD240 pKa = 5.41LMAPHH245 pKa = 6.54VLYY248 pKa = 11.1SHH250 pKa = 6.82ITSSTCSLGIAVLMLVRR267 pKa = 11.84WKK269 pKa = 8.92TAKK272 pKa = 9.32TRR274 pKa = 11.84ATVKK278 pKa = 10.56KK279 pKa = 10.59LLGSLLAVEE288 pKa = 4.86SGQMLLEE295 pKa = 4.4PPRR298 pKa = 11.84LRR300 pKa = 11.84NPAAAMFWYY309 pKa = 9.49KK310 pKa = 10.81KK311 pKa = 10.5AMCNSTVVTGEE322 pKa = 3.93TPQWILRR329 pKa = 11.84EE330 pKa = 3.98VSIQDD335 pKa = 3.49QVGEE339 pKa = 4.0QCLFSLSDD347 pKa = 3.67MVQWAYY353 pKa = 11.28DD354 pKa = 3.48NGIEE358 pKa = 4.23TEE360 pKa = 4.07SAAAYY365 pKa = 9.04EE366 pKa = 4.25YY367 pKa = 11.58ALLADD372 pKa = 4.03EE373 pKa = 5.67DD374 pKa = 4.38KK375 pKa = 11.52NADD378 pKa = 3.1AFLRR382 pKa = 11.84SNQQAKK388 pKa = 8.18WVGDD392 pKa = 3.59CMRR395 pKa = 11.84MVRR398 pKa = 11.84LYY400 pKa = 10.71RR401 pKa = 11.84RR402 pKa = 11.84AEE404 pKa = 4.03MNKK407 pKa = 7.28MTMGQWIKK415 pKa = 10.45HH416 pKa = 5.29RR417 pKa = 11.84SEE419 pKa = 3.94KK420 pKa = 9.39THH422 pKa = 7.36GEE424 pKa = 4.34GNWKK428 pKa = 10.38EE429 pKa = 3.63IFKK432 pKa = 10.4FLKK435 pKa = 10.04FQGIEE440 pKa = 3.59ITSFLTYY447 pKa = 10.17FRR449 pKa = 11.84LFLKK453 pKa = 10.23GVPKK457 pKa = 10.31KK458 pKa = 10.72NCIAICGPPNTGKK471 pKa = 10.57SLFGMSLIDD480 pKa = 4.62FIDD483 pKa = 3.73GRR485 pKa = 11.84VISHH489 pKa = 6.72CNSNSHH495 pKa = 6.72FWLQPLTEE503 pKa = 4.77CKK505 pKa = 10.13LALLDD510 pKa = 5.16DD511 pKa = 4.53ATPPTWDD518 pKa = 3.48YY519 pKa = 11.59FDD521 pKa = 3.58TYY523 pKa = 11.17LRR525 pKa = 11.84NLADD529 pKa = 4.28GNPVSVDD536 pKa = 3.3TKK538 pKa = 10.81HH539 pKa = 6.94RR540 pKa = 11.84APTQMKK546 pKa = 9.94CPPLLITTNTDD557 pKa = 2.23ISQGDD562 pKa = 3.07RR563 pKa = 11.84WTYY566 pKa = 9.23LKK568 pKa = 10.91SRR570 pKa = 11.84IKK572 pKa = 10.89VFTFPEE578 pKa = 4.07QMPLTEE584 pKa = 4.44MGDD587 pKa = 3.42PAFALTRR594 pKa = 11.84QNWKK598 pKa = 10.67SFFQRR603 pKa = 11.84CWSSLGLEE611 pKa = 4.51DD612 pKa = 5.6PEE614 pKa = 4.28QEE616 pKa = 4.56AANGDD621 pKa = 3.87PRR623 pKa = 11.84EE624 pKa = 4.0VLQPLRR630 pKa = 11.84CTARR634 pKa = 11.84RR635 pKa = 11.84ADD637 pKa = 3.65GLSS640 pKa = 3.1
Molecular weight: 71.44 kDa
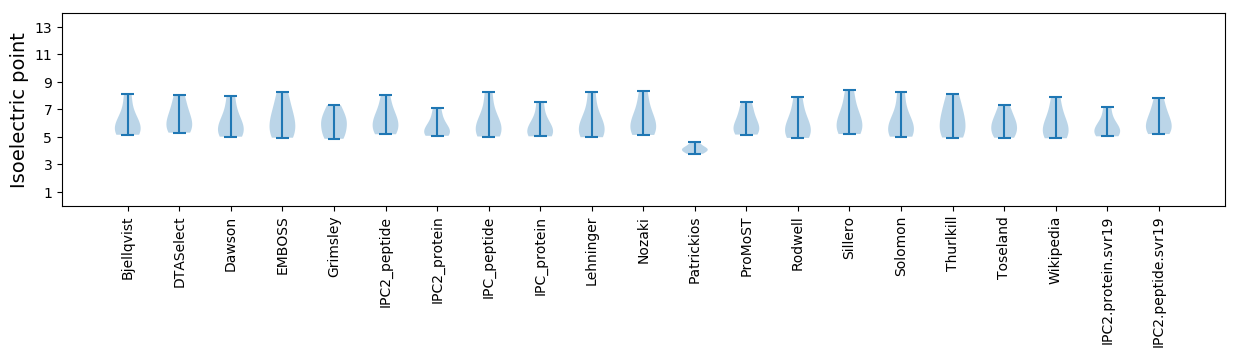
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2VIR0|F2VIR0_PSPV Minor capsid protein L2 OS=Phocoena phocoena papillomavirus 1 OX=706525 GN=L2 PE=3 SV=1
LL1 pKa = 7.21LHH3 pKa = 6.89LCLAPRR9 pKa = 11.84AVTSPLYY16 pKa = 10.43QLLEE20 pKa = 3.9EE21 pKa = 4.39TRR23 pKa = 11.84RR24 pKa = 11.84TRR26 pKa = 11.84QRR28 pKa = 11.84TSEE31 pKa = 4.02GHH33 pKa = 6.1PKK35 pKa = 10.23SGQGPMSAGILSQTQTQDD53 pKa = 3.11QSPTPQHH60 pKa = 6.51HH61 pKa = 5.97PHH63 pKa = 6.89ILCTPPRR70 pKa = 11.84SPTSSGSAATEE81 pKa = 4.06LTSPPPVSVSLTVGVPGAPEE101 pKa = 3.57ITLSFLLSS109 pKa = 3.46
LL1 pKa = 7.21LHH3 pKa = 6.89LCLAPRR9 pKa = 11.84AVTSPLYY16 pKa = 10.43QLLEE20 pKa = 3.9EE21 pKa = 4.39TRR23 pKa = 11.84RR24 pKa = 11.84TRR26 pKa = 11.84QRR28 pKa = 11.84TSEE31 pKa = 4.02GHH33 pKa = 6.1PKK35 pKa = 10.23SGQGPMSAGILSQTQTQDD53 pKa = 3.11QSPTPQHH60 pKa = 6.51HH61 pKa = 5.97PHH63 pKa = 6.89ILCTPPRR70 pKa = 11.84SPTSSGSAATEE81 pKa = 4.06LTSPPPVSVSLTVGVPGAPEE101 pKa = 3.57ITLSFLLSS109 pKa = 3.46
Molecular weight: 11.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2425 |
109 |
640 |
404.2 |
44.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.309 ± 0.429 | 2.474 ± 0.614 |
6.68 ± 0.461 | 5.526 ± 0.453 |
4.289 ± 0.339 | 7.918 ± 0.65 |
2.557 ± 0.405 | 3.546 ± 0.259 |
4.619 ± 0.619 | 8.412 ± 0.505 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.897 ± 0.317 | 2.763 ± 0.716 |
6.969 ± 0.98 | 3.835 ± 0.588 |
5.567 ± 0.212 | 8.742 ± 0.789 |
7.052 ± 0.423 | 6.639 ± 0.482 |
1.649 ± 0.353 | 2.557 ± 0.397 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
