
Thermodesulforhabdus norvegica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Syntrophobacterales; Syntrophobacteraceae; Thermodesulforhabdus
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
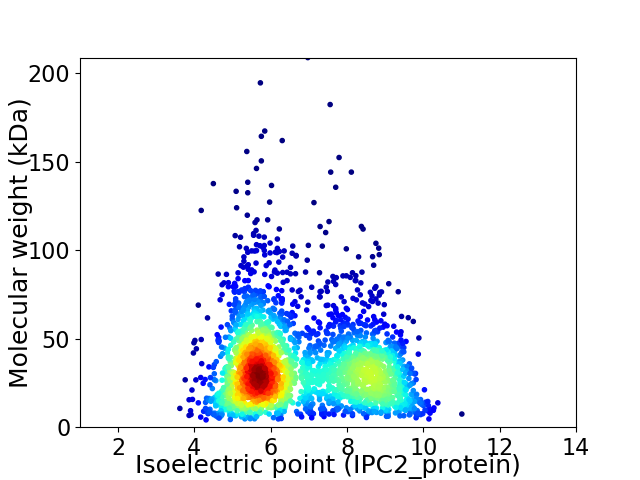
Virtual 2D-PAGE plot for 2661 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4SNQ6|A0A1I4SNQ6_9DELT Thiol:disulfide interchange protein OS=Thermodesulforhabdus norvegica OX=39841 GN=SAMN05660836_01076 PE=3 SV=1
MM1 pKa = 7.77RR2 pKa = 11.84KK3 pKa = 9.37YY4 pKa = 10.99LVMLIALVAAVAIAVPSFAVEE25 pKa = 4.36FKK27 pKa = 10.84YY28 pKa = 10.9GGMYY32 pKa = 9.76RR33 pKa = 11.84WRR35 pKa = 11.84IHH37 pKa = 6.64ANEE40 pKa = 3.85NMKK43 pKa = 10.43DD44 pKa = 3.68ANSDD48 pKa = 3.86LDD50 pKa = 4.57DD51 pKa = 3.74TANWIDD57 pKa = 3.35QRR59 pKa = 11.84LRR61 pKa = 11.84LYY63 pKa = 8.77FTFVGSEE70 pKa = 3.75NLQLVTKK77 pKa = 10.12WEE79 pKa = 4.25ADD81 pKa = 3.87TLWGLEE87 pKa = 4.09KK88 pKa = 10.75GAGRR92 pKa = 11.84HH93 pKa = 6.42GGGDD97 pKa = 3.09WGADD101 pKa = 3.11AVNLEE106 pKa = 4.25MKK108 pKa = 10.35NVYY111 pKa = 10.32LDD113 pKa = 3.81FMIPNTPVRR122 pKa = 11.84ALVGVQGLSYY132 pKa = 10.97LDD134 pKa = 3.35GWIVADD140 pKa = 4.05DD141 pKa = 3.85ASAFVLKK148 pKa = 9.52TAFDD152 pKa = 3.91PVKK155 pKa = 11.08VEE157 pKa = 4.08LGYY160 pKa = 10.43IAGVNSDD167 pKa = 4.14PVTDD171 pKa = 4.05YY172 pKa = 11.85NDD174 pKa = 3.21MDD176 pKa = 3.96DD177 pKa = 3.68WFLALKK183 pKa = 9.85YY184 pKa = 10.91AEE186 pKa = 4.9GPFNAAAVLFYY197 pKa = 10.69QYY199 pKa = 11.62AHH201 pKa = 7.22DD202 pKa = 4.95ADD204 pKa = 3.75YY205 pKa = 11.0SYY207 pKa = 10.97PNSIGYY213 pKa = 10.01GGTGAVDD220 pKa = 3.4KK221 pKa = 10.68EE222 pKa = 4.35DD223 pKa = 3.42NHH225 pKa = 7.98LVDD228 pKa = 5.38LGFSLGYY235 pKa = 10.8KK236 pKa = 9.4MDD238 pKa = 3.36WLGAKK243 pKa = 10.08VNFVKK248 pKa = 10.88NLGGYY253 pKa = 9.99DD254 pKa = 3.14IAGTDD259 pKa = 3.25EE260 pKa = 4.91DD261 pKa = 4.26EE262 pKa = 5.96DD263 pKa = 4.62YY264 pKa = 11.16EE265 pKa = 4.55GWMVEE270 pKa = 3.95AALDD274 pKa = 4.11FYY276 pKa = 10.96MGAFTFTLGGFWTSDD291 pKa = 3.15DD292 pKa = 3.52FAYY295 pKa = 9.77PAGRR299 pKa = 11.84SHH301 pKa = 6.42YY302 pKa = 8.2WAEE305 pKa = 3.83IAGLGTLDD313 pKa = 3.92VNVAGNDD320 pKa = 3.59WYY322 pKa = 10.02TSSYY326 pKa = 10.73GMGGGLPNRR335 pKa = 11.84GDD337 pKa = 3.83YY338 pKa = 10.93DD339 pKa = 4.24AGDD342 pKa = 3.75APRR345 pKa = 11.84NLWTIHH351 pKa = 6.99AGVAWQALDD360 pKa = 3.52TTKK363 pKa = 9.61VTFNYY368 pKa = 10.28YY369 pKa = 10.83YY370 pKa = 10.74LGTDD374 pKa = 3.7DD375 pKa = 6.14DD376 pKa = 4.41VLANEE381 pKa = 4.71ATDD384 pKa = 4.52EE385 pKa = 4.19YY386 pKa = 10.86DD387 pKa = 4.05DD388 pKa = 5.42SIGHH392 pKa = 6.23EE393 pKa = 4.13LDD395 pKa = 4.5LYY397 pKa = 10.56IDD399 pKa = 4.06HH400 pKa = 7.36KK401 pKa = 11.69LMDD404 pKa = 4.52GLTLRR409 pKa = 11.84LVGAYY414 pKa = 10.55LIGDD418 pKa = 4.11DD419 pKa = 4.18ALSTNEE425 pKa = 4.17EE426 pKa = 4.04DD427 pKa = 4.15DD428 pKa = 4.27NIYY431 pKa = 10.84EE432 pKa = 4.08VGARR436 pKa = 11.84LLWKK440 pKa = 10.18FF441 pKa = 3.6
MM1 pKa = 7.77RR2 pKa = 11.84KK3 pKa = 9.37YY4 pKa = 10.99LVMLIALVAAVAIAVPSFAVEE25 pKa = 4.36FKK27 pKa = 10.84YY28 pKa = 10.9GGMYY32 pKa = 9.76RR33 pKa = 11.84WRR35 pKa = 11.84IHH37 pKa = 6.64ANEE40 pKa = 3.85NMKK43 pKa = 10.43DD44 pKa = 3.68ANSDD48 pKa = 3.86LDD50 pKa = 4.57DD51 pKa = 3.74TANWIDD57 pKa = 3.35QRR59 pKa = 11.84LRR61 pKa = 11.84LYY63 pKa = 8.77FTFVGSEE70 pKa = 3.75NLQLVTKK77 pKa = 10.12WEE79 pKa = 4.25ADD81 pKa = 3.87TLWGLEE87 pKa = 4.09KK88 pKa = 10.75GAGRR92 pKa = 11.84HH93 pKa = 6.42GGGDD97 pKa = 3.09WGADD101 pKa = 3.11AVNLEE106 pKa = 4.25MKK108 pKa = 10.35NVYY111 pKa = 10.32LDD113 pKa = 3.81FMIPNTPVRR122 pKa = 11.84ALVGVQGLSYY132 pKa = 10.97LDD134 pKa = 3.35GWIVADD140 pKa = 4.05DD141 pKa = 3.85ASAFVLKK148 pKa = 9.52TAFDD152 pKa = 3.91PVKK155 pKa = 11.08VEE157 pKa = 4.08LGYY160 pKa = 10.43IAGVNSDD167 pKa = 4.14PVTDD171 pKa = 4.05YY172 pKa = 11.85NDD174 pKa = 3.21MDD176 pKa = 3.96DD177 pKa = 3.68WFLALKK183 pKa = 9.85YY184 pKa = 10.91AEE186 pKa = 4.9GPFNAAAVLFYY197 pKa = 10.69QYY199 pKa = 11.62AHH201 pKa = 7.22DD202 pKa = 4.95ADD204 pKa = 3.75YY205 pKa = 11.0SYY207 pKa = 10.97PNSIGYY213 pKa = 10.01GGTGAVDD220 pKa = 3.4KK221 pKa = 10.68EE222 pKa = 4.35DD223 pKa = 3.42NHH225 pKa = 7.98LVDD228 pKa = 5.38LGFSLGYY235 pKa = 10.8KK236 pKa = 9.4MDD238 pKa = 3.36WLGAKK243 pKa = 10.08VNFVKK248 pKa = 10.88NLGGYY253 pKa = 9.99DD254 pKa = 3.14IAGTDD259 pKa = 3.25EE260 pKa = 4.91DD261 pKa = 4.26EE262 pKa = 5.96DD263 pKa = 4.62YY264 pKa = 11.16EE265 pKa = 4.55GWMVEE270 pKa = 3.95AALDD274 pKa = 4.11FYY276 pKa = 10.96MGAFTFTLGGFWTSDD291 pKa = 3.15DD292 pKa = 3.52FAYY295 pKa = 9.77PAGRR299 pKa = 11.84SHH301 pKa = 6.42YY302 pKa = 8.2WAEE305 pKa = 3.83IAGLGTLDD313 pKa = 3.92VNVAGNDD320 pKa = 3.59WYY322 pKa = 10.02TSSYY326 pKa = 10.73GMGGGLPNRR335 pKa = 11.84GDD337 pKa = 3.83YY338 pKa = 10.93DD339 pKa = 4.24AGDD342 pKa = 3.75APRR345 pKa = 11.84NLWTIHH351 pKa = 6.99AGVAWQALDD360 pKa = 3.52TTKK363 pKa = 9.61VTFNYY368 pKa = 10.28YY369 pKa = 10.83YY370 pKa = 10.74LGTDD374 pKa = 3.7DD375 pKa = 6.14DD376 pKa = 4.41VLANEE381 pKa = 4.71ATDD384 pKa = 4.52EE385 pKa = 4.19YY386 pKa = 10.86DD387 pKa = 4.05DD388 pKa = 5.42SIGHH392 pKa = 6.23EE393 pKa = 4.13LDD395 pKa = 4.5LYY397 pKa = 10.56IDD399 pKa = 4.06HH400 pKa = 7.36KK401 pKa = 11.69LMDD404 pKa = 4.52GLTLRR409 pKa = 11.84LVGAYY414 pKa = 10.55LIGDD418 pKa = 4.11DD419 pKa = 4.18ALSTNEE425 pKa = 4.17EE426 pKa = 4.04DD427 pKa = 4.15DD428 pKa = 4.27NIYY431 pKa = 10.84EE432 pKa = 4.08VGARR436 pKa = 11.84LLWKK440 pKa = 10.18FF441 pKa = 3.6
Molecular weight: 48.97 kDa
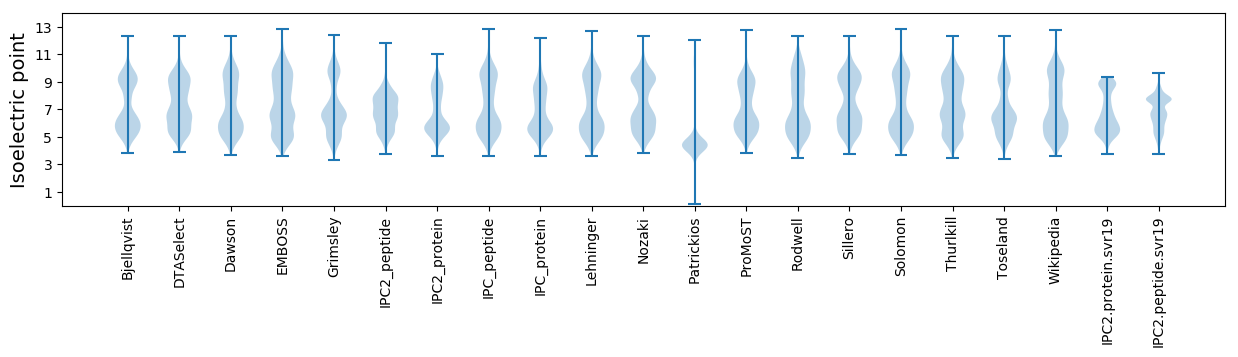
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4T512|A0A1I4T512_9DELT Pyruvate formate lyase activating enzyme OS=Thermodesulforhabdus norvegica OX=39841 GN=SAMN05660836_01234 PE=4 SV=1
MM1 pKa = 7.62AVPKK5 pKa = 10.38RR6 pKa = 11.84RR7 pKa = 11.84TSRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84NNRR16 pKa = 11.84RR17 pKa = 11.84SHH19 pKa = 6.59DD20 pKa = 4.73AISAPALYY28 pKa = 10.3LCPRR32 pKa = 11.84CKK34 pKa = 10.29SPKK37 pKa = 9.66LPHH40 pKa = 6.4RR41 pKa = 11.84VCPNCGTYY49 pKa = 10.04RR50 pKa = 11.84GRR52 pKa = 11.84DD53 pKa = 3.28VLKK56 pKa = 10.61IEE58 pKa = 4.42EE59 pKa = 4.2EE60 pKa = 4.13
MM1 pKa = 7.62AVPKK5 pKa = 10.38RR6 pKa = 11.84RR7 pKa = 11.84TSRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84NNRR16 pKa = 11.84RR17 pKa = 11.84SHH19 pKa = 6.59DD20 pKa = 4.73AISAPALYY28 pKa = 10.3LCPRR32 pKa = 11.84CKK34 pKa = 10.29SPKK37 pKa = 9.66LPHH40 pKa = 6.4RR41 pKa = 11.84VCPNCGTYY49 pKa = 10.04RR50 pKa = 11.84GRR52 pKa = 11.84DD53 pKa = 3.28VLKK56 pKa = 10.61IEE58 pKa = 4.42EE59 pKa = 4.2EE60 pKa = 4.13
Molecular weight: 6.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
865482 |
39 |
1866 |
325.2 |
36.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.719 ± 0.042 | 1.385 ± 0.021 |
5.031 ± 0.035 | 7.526 ± 0.049 |
4.409 ± 0.032 | 7.641 ± 0.041 |
1.979 ± 0.018 | 6.693 ± 0.042 |
5.361 ± 0.041 | 10.2 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.409 ± 0.02 | 2.94 ± 0.022 |
4.742 ± 0.027 | 2.543 ± 0.027 |
6.958 ± 0.041 | 5.577 ± 0.035 |
4.507 ± 0.027 | 7.991 ± 0.041 |
1.224 ± 0.02 | 3.166 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
