
Polaribacter reichenbachii
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
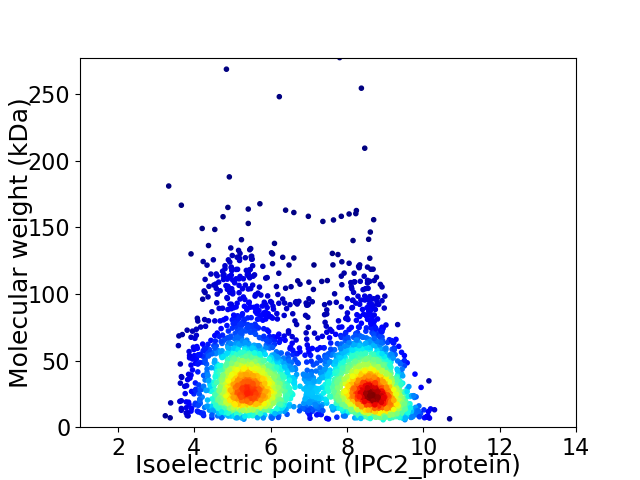
Virtual 2D-PAGE plot for 3418 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
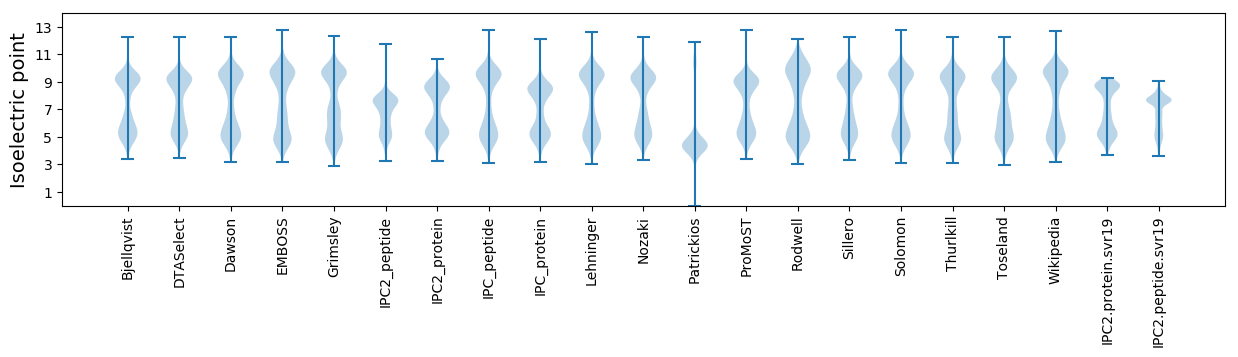
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B8U4M2|A0A1B8U4M2_9FLAO Peptidase_S8 domain-containing protein OS=Polaribacter reichenbachii OX=996801 GN=LPB301_05110 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.44SKK4 pKa = 10.85LSVFIFILFIFYY16 pKa = 10.82CKK18 pKa = 10.0DD19 pKa = 3.06VKK21 pKa = 11.06AQLSKK26 pKa = 11.05KK27 pKa = 10.04HH28 pKa = 6.45FIPPLTYY35 pKa = 10.53AEE37 pKa = 4.47EE38 pKa = 4.67GNANPEE44 pKa = 3.76SQYY47 pKa = 11.07FYY49 pKa = 10.92ISTPTNKK56 pKa = 9.95DD57 pKa = 2.52VSYY60 pKa = 9.75TIKK63 pKa = 10.78QIGQDD68 pKa = 3.32NDD70 pKa = 2.84ITGIVSSSNPQEE82 pKa = 3.95IFIATGDD89 pKa = 3.59SQLFVNANEE98 pKa = 4.26TGTVHH103 pKa = 5.92TNKK106 pKa = 10.34GYY108 pKa = 10.12IIEE111 pKa = 4.52ATDD114 pKa = 3.52VIYY117 pKa = 11.25VSVRR121 pKa = 11.84VLAGGGAQAGALVSKK136 pKa = 10.56GSSALGTTFRR146 pKa = 11.84AGMFTNEE153 pKa = 4.17NPQSNYY159 pKa = 10.6LNFVSVMASEE169 pKa = 5.52DD170 pKa = 3.41NTQITFDD177 pKa = 5.46DD178 pKa = 3.76ITPGLEE184 pKa = 3.57IKK186 pKa = 10.55NYY188 pKa = 9.26TSTFPFSVVLNEE200 pKa = 3.97GEE202 pKa = 4.38SYY204 pKa = 10.54IVATSADD211 pKa = 3.81DD212 pKa = 4.22VIVNRR217 pKa = 11.84DD218 pKa = 3.44GLIGTLISSDD228 pKa = 3.16KK229 pKa = 10.99NIVVNTGSANGSFHH243 pKa = 6.99NGGGRR248 pKa = 11.84DD249 pKa = 3.61YY250 pKa = 11.43GIDD253 pKa = 3.54QIVGADD259 pKa = 4.08KK260 pKa = 10.85IGTDD264 pKa = 3.82YY265 pKa = 11.29IFVKK269 pKa = 10.85GDD271 pKa = 3.51GSNGWEE277 pKa = 3.82NVLIVAHH284 pKa = 6.89EE285 pKa = 4.71DD286 pKa = 3.22NTVVSRR292 pKa = 11.84NGINSTINKK301 pKa = 9.71GEE303 pKa = 3.95YY304 pKa = 9.34TLIEE308 pKa = 4.2GDD310 pKa = 3.58EE311 pKa = 4.37FNANGNLFVKK321 pKa = 9.82TSKK324 pKa = 10.21PAFAYY329 pKa = 10.23QGVGANTSEE338 pKa = 4.34ANQGMFFVPPLSCEE352 pKa = 4.01SRR354 pKa = 11.84GKK356 pKa = 10.39VDD358 pKa = 4.6NIPNIEE364 pKa = 4.48SIGNTTFTGGITIVTNVNASVNINSQPITNFATSGPFSIDD404 pKa = 2.8IDD406 pKa = 3.75NDD408 pKa = 4.19GIADD412 pKa = 3.77YY413 pKa = 8.03EE414 pKa = 4.66TYY416 pKa = 10.77KK417 pKa = 9.7VTNLTGNISVDD428 pKa = 3.42SSDD431 pKa = 4.26EE432 pKa = 4.18LYY434 pKa = 10.82CAYY437 pKa = 10.75FNVNGAATSGSFYY450 pKa = 10.75SGFPTAPEE458 pKa = 4.06INFDD462 pKa = 3.93TTVSTLGNCIPNLTLEE478 pKa = 4.46AANTNLFDD486 pKa = 4.58SIKK489 pKa = 9.26WEE491 pKa = 4.32YY492 pKa = 10.66YY493 pKa = 10.79NEE495 pKa = 4.25TTSTWEE501 pKa = 3.84EE502 pKa = 3.48RR503 pKa = 11.84SSNNTYY509 pKa = 10.71KK510 pKa = 10.63PIEE513 pKa = 4.37SEE515 pKa = 3.69PGKK518 pKa = 10.63YY519 pKa = 9.73RR520 pKa = 11.84LIGTIDD526 pKa = 3.35CTGATFEE533 pKa = 4.72SIEE536 pKa = 4.36IPVSICPDD544 pKa = 3.77DD545 pKa = 4.31FDD547 pKa = 6.96GDD549 pKa = 4.63LIIDD553 pKa = 4.18NLDD556 pKa = 3.02VDD558 pKa = 4.21IDD560 pKa = 3.72NDD562 pKa = 4.77GILNCDD568 pKa = 3.39EE569 pKa = 4.47SLGNATLNLADD580 pKa = 4.8ADD582 pKa = 3.78NPEE585 pKa = 5.31IIFQDD590 pKa = 3.26SSTNSAILSRR600 pKa = 11.84VYY602 pKa = 10.8SEE604 pKa = 4.89SDD606 pKa = 3.17VTNTYY611 pKa = 9.23TGQNNGNFQSIIYY624 pKa = 9.21PSVDD628 pKa = 2.7SKK630 pKa = 11.46LQYY633 pKa = 10.44EE634 pKa = 4.64LNFTQNVNFKK644 pKa = 8.89FTQNRR649 pKa = 11.84NLDD652 pKa = 3.53HH653 pKa = 6.94TISDD657 pKa = 3.98GEE659 pKa = 4.14FFILKK664 pKa = 10.04IGPSNKK670 pKa = 9.51NVTLLDD676 pKa = 4.56PDD678 pKa = 4.11DD679 pKa = 4.01QLLIDD684 pKa = 4.45SNFDD688 pKa = 3.46GEE690 pKa = 4.52FEE692 pKa = 4.32SGITNISASEE702 pKa = 3.48IHH704 pKa = 6.63FKK706 pKa = 11.17YY707 pKa = 10.25KK708 pKa = 10.99ANTTGASSNFKK719 pKa = 10.26FVANQVDD726 pKa = 3.9QIDD729 pKa = 4.42FKK731 pKa = 11.29HH732 pKa = 6.07QSTGLAASSTFSGNLEE748 pKa = 4.19LTCFTLDD755 pKa = 3.25SDD757 pKa = 4.36GDD759 pKa = 4.15GIEE762 pKa = 4.59NMFDD766 pKa = 3.83LDD768 pKa = 4.16SDD770 pKa = 3.87NDD772 pKa = 4.8GIPDD776 pKa = 3.4ISEE779 pKa = 3.91SSATKK784 pKa = 10.21IILSNTDD791 pKa = 3.0ADD793 pKa = 5.33LDD795 pKa = 3.92GLDD798 pKa = 4.53DD799 pKa = 3.93VFIGIITNIDD809 pKa = 3.4TDD811 pKa = 3.58NDD813 pKa = 4.82GIPNHH818 pKa = 6.95LDD820 pKa = 2.75VDD822 pKa = 4.04ADD824 pKa = 3.52NDD826 pKa = 4.82GIFDD830 pKa = 3.89FVEE833 pKa = 4.17AGITEE838 pKa = 4.16TDD840 pKa = 3.41ANNDD844 pKa = 4.17GILDD848 pKa = 4.19DD849 pKa = 4.13ATTANVGLNGLLNALEE865 pKa = 4.22TSPDD869 pKa = 3.73NNILAYY875 pKa = 9.65TILDD879 pKa = 3.5TDD881 pKa = 4.1ADD883 pKa = 3.94NIYY886 pKa = 11.24NFLEE890 pKa = 4.31LDD892 pKa = 3.94ADD894 pKa = 3.83NDD896 pKa = 3.5NCFDD900 pKa = 3.57VFEE903 pKa = 6.32AGFTDD908 pKa = 4.12NNDD911 pKa = 3.86DD912 pKa = 4.9GILDD916 pKa = 3.73ATPFSVNNNGKK927 pKa = 9.94IINNSDD933 pKa = 3.49GYY935 pKa = 6.96TTPNSDD941 pKa = 4.72YY942 pKa = 9.7ITSAPIEE949 pKa = 4.27INTPFEE955 pKa = 4.41DD956 pKa = 5.37VIFCEE961 pKa = 4.06NDD963 pKa = 2.86TDD965 pKa = 4.84VITIDD970 pKa = 3.3TTADD974 pKa = 3.13SFQWQISPDD983 pKa = 3.31GSTWSSITDD992 pKa = 3.19NAVYY996 pKa = 10.75SNSTSASLQITNTPLSYY1013 pKa = 10.69HH1014 pKa = 6.45NYY1016 pKa = 7.99QYY1018 pKa = 11.21RR1019 pKa = 11.84VILNRR1024 pKa = 11.84TGNACTEE1031 pKa = 4.19TSNSITLTVNPLPILKK1047 pKa = 10.33ANPEE1051 pKa = 4.31LNQCIEE1057 pKa = 4.15VADD1060 pKa = 4.14TNTTVNLTLAEE1071 pKa = 4.21INISEE1076 pKa = 4.45TTNVTFEE1083 pKa = 4.46YY1084 pKa = 10.4YY1085 pKa = 10.75EE1086 pKa = 4.46DD1087 pKa = 3.45MAGANLIADD1096 pKa = 3.85PTSYY1100 pKa = 10.29PVQVNITEE1108 pKa = 4.25TVYY1111 pKa = 11.2VKK1113 pKa = 10.69VISEE1117 pKa = 4.3FSCEE1121 pKa = 3.89TDD1123 pKa = 4.02LSEE1126 pKa = 4.16LTINVGQTIRR1136 pKa = 11.84NSYY1139 pKa = 11.08NDD1141 pKa = 4.28LQPPVCDD1148 pKa = 4.84DD1149 pKa = 3.9FLDD1152 pKa = 3.9VDD1154 pKa = 4.34GNDD1157 pKa = 3.37TAVNSDD1163 pKa = 3.38TDD1165 pKa = 3.98NITNFYY1171 pKa = 10.25LDD1173 pKa = 3.22KK1174 pKa = 10.43TAIINGINPPMNTEE1188 pKa = 3.7VFFYY1192 pKa = 10.61EE1193 pKa = 4.47NEE1195 pKa = 4.0SDD1197 pKa = 4.37RR1198 pKa = 11.84DD1199 pKa = 3.6NSLNEE1204 pKa = 3.52IDD1206 pKa = 3.64VTNFRR1211 pKa = 11.84NNISKK1216 pKa = 10.81NEE1218 pKa = 4.09VINIPNGIQFPIFYY1232 pKa = 10.01KK1233 pKa = 10.42ILSTINNDD1241 pKa = 3.36CQGIGEE1247 pKa = 4.85FYY1249 pKa = 10.84VQIDD1253 pKa = 4.01TVPTAEE1259 pKa = 4.19VVPDD1263 pKa = 5.27LEE1265 pKa = 5.18LCDD1268 pKa = 5.09DD1269 pKa = 5.4AIDD1272 pKa = 4.19GNATNGIVQYY1282 pKa = 11.0FDD1284 pKa = 5.42LEE1286 pKa = 4.47SQTTAILNGQNAADD1300 pKa = 3.87FTVTYY1305 pKa = 10.2HH1306 pKa = 7.98DD1307 pKa = 4.73SEE1309 pKa = 4.4TDD1311 pKa = 3.15ANAGIDD1317 pKa = 3.71EE1318 pKa = 4.34LTSPFEE1324 pKa = 3.78NSVRR1328 pKa = 11.84DD1329 pKa = 3.63SQPIYY1334 pKa = 11.18VRR1336 pKa = 11.84VTNNTTGCFTDD1347 pKa = 3.2HH1348 pKa = 6.83TSFNVIVNPVPIANFVNDD1366 pKa = 5.67LEE1368 pKa = 4.59VCDD1371 pKa = 5.04DD1372 pKa = 4.13NSDD1375 pKa = 3.19GSARR1379 pKa = 11.84NGFSQSIDD1387 pKa = 3.94LEE1389 pKa = 4.39SQTAGILGAQDD1400 pKa = 3.75PNIHH1404 pKa = 6.17IVTYY1408 pKa = 10.27HH1409 pKa = 7.04RR1410 pKa = 11.84SLADD1414 pKa = 3.47AQSGNLPLVSPYY1426 pKa = 10.63TNNTIDD1432 pKa = 3.64RR1433 pKa = 11.84EE1434 pKa = 4.41TIYY1437 pKa = 11.16VRR1439 pKa = 11.84ILNTNSMCVNGISNFDD1455 pKa = 3.86VIVNPEE1461 pKa = 4.09PTFEE1465 pKa = 4.35TPTNLAYY1472 pKa = 10.77CDD1474 pKa = 4.77DD1475 pKa = 5.16DD1476 pKa = 6.7LDD1478 pKa = 6.17GDD1480 pKa = 4.13DD1481 pKa = 5.54ANGIVQNIDD1490 pKa = 3.41LDD1492 pKa = 4.27SKK1494 pKa = 9.78ITEE1497 pKa = 4.09ILGASQSPNDD1507 pKa = 3.63FWVTFHH1513 pKa = 7.59KK1514 pKa = 10.75SQADD1518 pKa = 3.5ATSGNDD1524 pKa = 3.51PVVSPYY1530 pKa = 11.15QNTT1533 pKa = 3.29
MM1 pKa = 7.49KK2 pKa = 10.44SKK4 pKa = 10.85LSVFIFILFIFYY16 pKa = 10.82CKK18 pKa = 10.0DD19 pKa = 3.06VKK21 pKa = 11.06AQLSKK26 pKa = 11.05KK27 pKa = 10.04HH28 pKa = 6.45FIPPLTYY35 pKa = 10.53AEE37 pKa = 4.47EE38 pKa = 4.67GNANPEE44 pKa = 3.76SQYY47 pKa = 11.07FYY49 pKa = 10.92ISTPTNKK56 pKa = 9.95DD57 pKa = 2.52VSYY60 pKa = 9.75TIKK63 pKa = 10.78QIGQDD68 pKa = 3.32NDD70 pKa = 2.84ITGIVSSSNPQEE82 pKa = 3.95IFIATGDD89 pKa = 3.59SQLFVNANEE98 pKa = 4.26TGTVHH103 pKa = 5.92TNKK106 pKa = 10.34GYY108 pKa = 10.12IIEE111 pKa = 4.52ATDD114 pKa = 3.52VIYY117 pKa = 11.25VSVRR121 pKa = 11.84VLAGGGAQAGALVSKK136 pKa = 10.56GSSALGTTFRR146 pKa = 11.84AGMFTNEE153 pKa = 4.17NPQSNYY159 pKa = 10.6LNFVSVMASEE169 pKa = 5.52DD170 pKa = 3.41NTQITFDD177 pKa = 5.46DD178 pKa = 3.76ITPGLEE184 pKa = 3.57IKK186 pKa = 10.55NYY188 pKa = 9.26TSTFPFSVVLNEE200 pKa = 3.97GEE202 pKa = 4.38SYY204 pKa = 10.54IVATSADD211 pKa = 3.81DD212 pKa = 4.22VIVNRR217 pKa = 11.84DD218 pKa = 3.44GLIGTLISSDD228 pKa = 3.16KK229 pKa = 10.99NIVVNTGSANGSFHH243 pKa = 6.99NGGGRR248 pKa = 11.84DD249 pKa = 3.61YY250 pKa = 11.43GIDD253 pKa = 3.54QIVGADD259 pKa = 4.08KK260 pKa = 10.85IGTDD264 pKa = 3.82YY265 pKa = 11.29IFVKK269 pKa = 10.85GDD271 pKa = 3.51GSNGWEE277 pKa = 3.82NVLIVAHH284 pKa = 6.89EE285 pKa = 4.71DD286 pKa = 3.22NTVVSRR292 pKa = 11.84NGINSTINKK301 pKa = 9.71GEE303 pKa = 3.95YY304 pKa = 9.34TLIEE308 pKa = 4.2GDD310 pKa = 3.58EE311 pKa = 4.37FNANGNLFVKK321 pKa = 9.82TSKK324 pKa = 10.21PAFAYY329 pKa = 10.23QGVGANTSEE338 pKa = 4.34ANQGMFFVPPLSCEE352 pKa = 4.01SRR354 pKa = 11.84GKK356 pKa = 10.39VDD358 pKa = 4.6NIPNIEE364 pKa = 4.48SIGNTTFTGGITIVTNVNASVNINSQPITNFATSGPFSIDD404 pKa = 2.8IDD406 pKa = 3.75NDD408 pKa = 4.19GIADD412 pKa = 3.77YY413 pKa = 8.03EE414 pKa = 4.66TYY416 pKa = 10.77KK417 pKa = 9.7VTNLTGNISVDD428 pKa = 3.42SSDD431 pKa = 4.26EE432 pKa = 4.18LYY434 pKa = 10.82CAYY437 pKa = 10.75FNVNGAATSGSFYY450 pKa = 10.75SGFPTAPEE458 pKa = 4.06INFDD462 pKa = 3.93TTVSTLGNCIPNLTLEE478 pKa = 4.46AANTNLFDD486 pKa = 4.58SIKK489 pKa = 9.26WEE491 pKa = 4.32YY492 pKa = 10.66YY493 pKa = 10.79NEE495 pKa = 4.25TTSTWEE501 pKa = 3.84EE502 pKa = 3.48RR503 pKa = 11.84SSNNTYY509 pKa = 10.71KK510 pKa = 10.63PIEE513 pKa = 4.37SEE515 pKa = 3.69PGKK518 pKa = 10.63YY519 pKa = 9.73RR520 pKa = 11.84LIGTIDD526 pKa = 3.35CTGATFEE533 pKa = 4.72SIEE536 pKa = 4.36IPVSICPDD544 pKa = 3.77DD545 pKa = 4.31FDD547 pKa = 6.96GDD549 pKa = 4.63LIIDD553 pKa = 4.18NLDD556 pKa = 3.02VDD558 pKa = 4.21IDD560 pKa = 3.72NDD562 pKa = 4.77GILNCDD568 pKa = 3.39EE569 pKa = 4.47SLGNATLNLADD580 pKa = 4.8ADD582 pKa = 3.78NPEE585 pKa = 5.31IIFQDD590 pKa = 3.26SSTNSAILSRR600 pKa = 11.84VYY602 pKa = 10.8SEE604 pKa = 4.89SDD606 pKa = 3.17VTNTYY611 pKa = 9.23TGQNNGNFQSIIYY624 pKa = 9.21PSVDD628 pKa = 2.7SKK630 pKa = 11.46LQYY633 pKa = 10.44EE634 pKa = 4.64LNFTQNVNFKK644 pKa = 8.89FTQNRR649 pKa = 11.84NLDD652 pKa = 3.53HH653 pKa = 6.94TISDD657 pKa = 3.98GEE659 pKa = 4.14FFILKK664 pKa = 10.04IGPSNKK670 pKa = 9.51NVTLLDD676 pKa = 4.56PDD678 pKa = 4.11DD679 pKa = 4.01QLLIDD684 pKa = 4.45SNFDD688 pKa = 3.46GEE690 pKa = 4.52FEE692 pKa = 4.32SGITNISASEE702 pKa = 3.48IHH704 pKa = 6.63FKK706 pKa = 11.17YY707 pKa = 10.25KK708 pKa = 10.99ANTTGASSNFKK719 pKa = 10.26FVANQVDD726 pKa = 3.9QIDD729 pKa = 4.42FKK731 pKa = 11.29HH732 pKa = 6.07QSTGLAASSTFSGNLEE748 pKa = 4.19LTCFTLDD755 pKa = 3.25SDD757 pKa = 4.36GDD759 pKa = 4.15GIEE762 pKa = 4.59NMFDD766 pKa = 3.83LDD768 pKa = 4.16SDD770 pKa = 3.87NDD772 pKa = 4.8GIPDD776 pKa = 3.4ISEE779 pKa = 3.91SSATKK784 pKa = 10.21IILSNTDD791 pKa = 3.0ADD793 pKa = 5.33LDD795 pKa = 3.92GLDD798 pKa = 4.53DD799 pKa = 3.93VFIGIITNIDD809 pKa = 3.4TDD811 pKa = 3.58NDD813 pKa = 4.82GIPNHH818 pKa = 6.95LDD820 pKa = 2.75VDD822 pKa = 4.04ADD824 pKa = 3.52NDD826 pKa = 4.82GIFDD830 pKa = 3.89FVEE833 pKa = 4.17AGITEE838 pKa = 4.16TDD840 pKa = 3.41ANNDD844 pKa = 4.17GILDD848 pKa = 4.19DD849 pKa = 4.13ATTANVGLNGLLNALEE865 pKa = 4.22TSPDD869 pKa = 3.73NNILAYY875 pKa = 9.65TILDD879 pKa = 3.5TDD881 pKa = 4.1ADD883 pKa = 3.94NIYY886 pKa = 11.24NFLEE890 pKa = 4.31LDD892 pKa = 3.94ADD894 pKa = 3.83NDD896 pKa = 3.5NCFDD900 pKa = 3.57VFEE903 pKa = 6.32AGFTDD908 pKa = 4.12NNDD911 pKa = 3.86DD912 pKa = 4.9GILDD916 pKa = 3.73ATPFSVNNNGKK927 pKa = 9.94IINNSDD933 pKa = 3.49GYY935 pKa = 6.96TTPNSDD941 pKa = 4.72YY942 pKa = 9.7ITSAPIEE949 pKa = 4.27INTPFEE955 pKa = 4.41DD956 pKa = 5.37VIFCEE961 pKa = 4.06NDD963 pKa = 2.86TDD965 pKa = 4.84VITIDD970 pKa = 3.3TTADD974 pKa = 3.13SFQWQISPDD983 pKa = 3.31GSTWSSITDD992 pKa = 3.19NAVYY996 pKa = 10.75SNSTSASLQITNTPLSYY1013 pKa = 10.69HH1014 pKa = 6.45NYY1016 pKa = 7.99QYY1018 pKa = 11.21RR1019 pKa = 11.84VILNRR1024 pKa = 11.84TGNACTEE1031 pKa = 4.19TSNSITLTVNPLPILKK1047 pKa = 10.33ANPEE1051 pKa = 4.31LNQCIEE1057 pKa = 4.15VADD1060 pKa = 4.14TNTTVNLTLAEE1071 pKa = 4.21INISEE1076 pKa = 4.45TTNVTFEE1083 pKa = 4.46YY1084 pKa = 10.4YY1085 pKa = 10.75EE1086 pKa = 4.46DD1087 pKa = 3.45MAGANLIADD1096 pKa = 3.85PTSYY1100 pKa = 10.29PVQVNITEE1108 pKa = 4.25TVYY1111 pKa = 11.2VKK1113 pKa = 10.69VISEE1117 pKa = 4.3FSCEE1121 pKa = 3.89TDD1123 pKa = 4.02LSEE1126 pKa = 4.16LTINVGQTIRR1136 pKa = 11.84NSYY1139 pKa = 11.08NDD1141 pKa = 4.28LQPPVCDD1148 pKa = 4.84DD1149 pKa = 3.9FLDD1152 pKa = 3.9VDD1154 pKa = 4.34GNDD1157 pKa = 3.37TAVNSDD1163 pKa = 3.38TDD1165 pKa = 3.98NITNFYY1171 pKa = 10.25LDD1173 pKa = 3.22KK1174 pKa = 10.43TAIINGINPPMNTEE1188 pKa = 3.7VFFYY1192 pKa = 10.61EE1193 pKa = 4.47NEE1195 pKa = 4.0SDD1197 pKa = 4.37RR1198 pKa = 11.84DD1199 pKa = 3.6NSLNEE1204 pKa = 3.52IDD1206 pKa = 3.64VTNFRR1211 pKa = 11.84NNISKK1216 pKa = 10.81NEE1218 pKa = 4.09VINIPNGIQFPIFYY1232 pKa = 10.01KK1233 pKa = 10.42ILSTINNDD1241 pKa = 3.36CQGIGEE1247 pKa = 4.85FYY1249 pKa = 10.84VQIDD1253 pKa = 4.01TVPTAEE1259 pKa = 4.19VVPDD1263 pKa = 5.27LEE1265 pKa = 5.18LCDD1268 pKa = 5.09DD1269 pKa = 5.4AIDD1272 pKa = 4.19GNATNGIVQYY1282 pKa = 11.0FDD1284 pKa = 5.42LEE1286 pKa = 4.47SQTTAILNGQNAADD1300 pKa = 3.87FTVTYY1305 pKa = 10.2HH1306 pKa = 7.98DD1307 pKa = 4.73SEE1309 pKa = 4.4TDD1311 pKa = 3.15ANAGIDD1317 pKa = 3.71EE1318 pKa = 4.34LTSPFEE1324 pKa = 3.78NSVRR1328 pKa = 11.84DD1329 pKa = 3.63SQPIYY1334 pKa = 11.18VRR1336 pKa = 11.84VTNNTTGCFTDD1347 pKa = 3.2HH1348 pKa = 6.83TSFNVIVNPVPIANFVNDD1366 pKa = 5.67LEE1368 pKa = 4.59VCDD1371 pKa = 5.04DD1372 pKa = 4.13NSDD1375 pKa = 3.19GSARR1379 pKa = 11.84NGFSQSIDD1387 pKa = 3.94LEE1389 pKa = 4.39SQTAGILGAQDD1400 pKa = 3.75PNIHH1404 pKa = 6.17IVTYY1408 pKa = 10.27HH1409 pKa = 7.04RR1410 pKa = 11.84SLADD1414 pKa = 3.47AQSGNLPLVSPYY1426 pKa = 10.63TNNTIDD1432 pKa = 3.64RR1433 pKa = 11.84EE1434 pKa = 4.41TIYY1437 pKa = 11.16VRR1439 pKa = 11.84ILNTNSMCVNGISNFDD1455 pKa = 3.86VIVNPEE1461 pKa = 4.09PTFEE1465 pKa = 4.35TPTNLAYY1472 pKa = 10.77CDD1474 pKa = 4.77DD1475 pKa = 5.16DD1476 pKa = 6.7LDD1478 pKa = 6.17GDD1480 pKa = 4.13DD1481 pKa = 5.54ANGIVQNIDD1490 pKa = 3.41LDD1492 pKa = 4.27SKK1494 pKa = 9.78ITEE1497 pKa = 4.09ILGASQSPNDD1507 pKa = 3.63FWVTFHH1513 pKa = 7.59KK1514 pKa = 10.75SQADD1518 pKa = 3.5ATSGNDD1524 pKa = 3.51PVVSPYY1530 pKa = 11.15QNTT1533 pKa = 3.29
Molecular weight: 166.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B8TWH8|A0A1B8TWH8_9FLAO Malate dehydrogenase OS=Polaribacter reichenbachii OX=996801 GN=mdh PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 3.92TITTDD27 pKa = 3.73KK28 pKa = 10.91PEE30 pKa = 4.37KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.38RR39 pKa = 11.84SGGRR43 pKa = 11.84NNQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.99KK59 pKa = 9.43QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.84FKK68 pKa = 10.95RR69 pKa = 11.84DD70 pKa = 3.68KK71 pKa = 11.21NDD73 pKa = 2.9MPATVKK79 pKa = 10.14TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALLSYY96 pKa = 11.49ADD98 pKa = 3.5GEE100 pKa = 4.22KK101 pKa = 10.25RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.19VGQTIVSGSGVSPEE125 pKa = 4.69IGNAMPLSEE134 pKa = 4.97IPLGTTISCIEE145 pKa = 3.88LHH147 pKa = 6.66PGQGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILLTCSATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 8.88AGRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.4ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 3.92TITTDD27 pKa = 3.73KK28 pKa = 10.91PEE30 pKa = 4.37KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.38RR39 pKa = 11.84SGGRR43 pKa = 11.84NNQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.99KK59 pKa = 9.43QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.84FKK68 pKa = 10.95RR69 pKa = 11.84DD70 pKa = 3.68KK71 pKa = 11.21NDD73 pKa = 2.9MPATVKK79 pKa = 10.14TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALLSYY96 pKa = 11.49ADD98 pKa = 3.5GEE100 pKa = 4.22KK101 pKa = 10.25RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.19VGQTIVSGSGVSPEE125 pKa = 4.69IGNAMPLSEE134 pKa = 4.97IPLGTTISCIEE145 pKa = 3.88LHH147 pKa = 6.66PGQGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILLTCSATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 8.88AGRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.4ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 29.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1227233 |
50 |
2383 |
359.1 |
40.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.955 ± 0.046 | 0.656 ± 0.011 |
5.61 ± 0.029 | 6.479 ± 0.041 |
5.46 ± 0.041 | 6.142 ± 0.044 |
1.624 ± 0.018 | 8.363 ± 0.04 |
8.775 ± 0.056 | 9.038 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.948 ± 0.019 | 6.952 ± 0.051 |
3.13 ± 0.02 | 3.156 ± 0.022 |
3.085 ± 0.022 | 6.57 ± 0.033 |
5.857 ± 0.034 | 5.967 ± 0.027 |
1.115 ± 0.016 | 4.12 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
