
Wenzhou weivirus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.33
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KL43|A0A1L3KL43_9VIRU RNA-dependent RNA polymerase OS=Wenzhou weivirus-like virus 1 OX=1923682 PE=4 SV=1
MM1 pKa = 7.36LSVRR5 pKa = 11.84EE6 pKa = 4.23GFSDD10 pKa = 3.92LKK12 pKa = 10.33YY13 pKa = 11.25GEE15 pKa = 4.17VAPYY19 pKa = 10.24LRR21 pKa = 11.84EE22 pKa = 3.83RR23 pKa = 11.84AAKK26 pKa = 9.99RR27 pKa = 11.84QGEE30 pKa = 4.27WTLTVHH36 pKa = 6.0EE37 pKa = 5.67AEE39 pKa = 4.93DD40 pKa = 3.44SWLGILSRR48 pKa = 11.84AVSFKK53 pKa = 10.98VVLTEE58 pKa = 3.73EE59 pKa = 4.88EE60 pKa = 4.31YY61 pKa = 11.4AKK63 pKa = 10.67LSRR66 pKa = 11.84ALYY69 pKa = 9.61HH70 pKa = 7.03GIRR73 pKa = 11.84NGTKK77 pKa = 8.8KK78 pKa = 9.31TSLLANLEE86 pKa = 4.03NTAQALWPTGSLDD99 pKa = 3.55HH100 pKa = 6.81VAMRR104 pKa = 11.84HH105 pKa = 4.8LAVTAVVQYY114 pKa = 7.36VTKK117 pKa = 10.44EE118 pKa = 3.77STPQGFRR125 pKa = 11.84LPYY128 pKa = 8.31WMTWGAMVTQATVGVHH144 pKa = 4.45ILGRR148 pKa = 11.84LVKK151 pKa = 10.35GAARR155 pKa = 11.84RR156 pKa = 11.84AWITGPHH163 pKa = 6.99PGPFGQLIMRR173 pKa = 11.84EE174 pKa = 4.28YY175 pKa = 10.32FATPALVMWALMTVISYY192 pKa = 10.52CGARR196 pKa = 11.84GITLSWRR203 pKa = 11.84SVLPVLSRR211 pKa = 11.84NAGTTRR217 pKa = 11.84TGPAAEE223 pKa = 4.72AAPAAPGGPPPAGATPGAGGAAPGPAGGAPPSGGTPAAGAPAPPSPPVMEE273 pKa = 4.43QPATPEE279 pKa = 4.12DD280 pKa = 4.01PEE282 pKa = 4.09NAANASEE289 pKa = 4.39DD290 pKa = 3.81LRR292 pKa = 11.84TMEE295 pKa = 4.38MPGKK299 pKa = 9.9IVGVLGQNFDD309 pKa = 3.24KK310 pKa = 10.92DD311 pKa = 3.91LPVNHH316 pKa = 6.78LPVVGVLVGPCQRR329 pKa = 11.84KK330 pKa = 8.28PNVYY334 pKa = 10.3SKK336 pKa = 9.71TVEE339 pKa = 4.0NLKK342 pKa = 10.56AAINEE347 pKa = 4.26RR348 pKa = 11.84IIKK351 pKa = 9.84KK352 pKa = 9.6ARR354 pKa = 11.84KK355 pKa = 8.64PKK357 pKa = 9.95LCRR360 pKa = 11.84ADD362 pKa = 3.37RR363 pKa = 11.84VRR365 pKa = 11.84ISRR368 pKa = 11.84LVRR371 pKa = 11.84KK372 pKa = 9.96AMSNSEE378 pKa = 3.84KK379 pKa = 10.61HH380 pKa = 6.06GVFSKK385 pKa = 10.94SRR387 pKa = 11.84IQEE390 pKa = 3.46WAIQHH395 pKa = 5.96FNLEE399 pKa = 4.26EE400 pKa = 4.01MKK402 pKa = 10.26SQKK405 pKa = 9.45WSLEE409 pKa = 3.81RR410 pKa = 11.84FRR412 pKa = 11.84NALKK416 pKa = 10.58NLYY419 pKa = 9.78EE420 pKa = 4.65KK421 pKa = 11.14DD422 pKa = 3.47PVEE425 pKa = 4.51FMLKK429 pKa = 10.47AGIKK433 pKa = 8.23PEE435 pKa = 3.94CMEE438 pKa = 4.21EE439 pKa = 4.29GKK441 pKa = 10.61APRR444 pKa = 11.84MLIADD449 pKa = 3.91GDD451 pKa = 4.11EE452 pKa = 4.41GQLMALAVIKK462 pKa = 10.61CFEE465 pKa = 4.11EE466 pKa = 4.39LLFKK470 pKa = 10.82HH471 pKa = 6.16FEE473 pKa = 3.99EE474 pKa = 5.51RR475 pKa = 11.84SIKK478 pKa = 10.33HH479 pKa = 5.45LSKK482 pKa = 10.65HH483 pKa = 4.81EE484 pKa = 3.85AMEE487 pKa = 4.0RR488 pKa = 11.84VVRR491 pKa = 11.84VLKK494 pKa = 10.84KK495 pKa = 9.43KK496 pKa = 9.74GARR499 pKa = 11.84AVEE502 pKa = 4.36GDD504 pKa = 3.23GSAWDD509 pKa = 3.99TTCGVLVRR517 pKa = 11.84SLIEE521 pKa = 3.88NPILFHH527 pKa = 7.03IFRR530 pKa = 11.84TLAEE534 pKa = 4.3FGVIPEE540 pKa = 3.86TWMAEE545 pKa = 3.64HH546 pKa = 6.86HH547 pKa = 6.15AACTKK552 pKa = 10.58EE553 pKa = 3.72KK554 pKa = 10.95LKK556 pKa = 11.22LFFSNKK562 pKa = 6.27FQKK565 pKa = 9.2VTVTIDD571 pKa = 3.78AIRR574 pKa = 11.84RR575 pKa = 11.84SGHH578 pKa = 6.55RR579 pKa = 11.84GTSCLNWWINFTMWVSSIFKK599 pKa = 9.87EE600 pKa = 4.25PEE602 pKa = 3.66RR603 pKa = 11.84FLDD606 pKa = 3.49PDD608 pKa = 3.5VRR610 pKa = 11.84KK611 pKa = 8.86GTDD614 pKa = 3.13LTEE617 pKa = 4.62KK618 pKa = 9.66EE619 pKa = 4.24RR620 pKa = 11.84WWNGTFEE627 pKa = 5.69GDD629 pKa = 4.45DD630 pKa = 4.39SLCTMCPPMLKK641 pKa = 9.7DD642 pKa = 4.93DD643 pKa = 3.87KK644 pKa = 10.92MSDD647 pKa = 3.23IFCQFWSDD655 pKa = 2.97AGFNMKK661 pKa = 9.61IVYY664 pKa = 9.59CDD666 pKa = 3.13RR667 pKa = 11.84RR668 pKa = 11.84ATFTGWHH675 pKa = 5.94LVCEE679 pKa = 4.38NGEE682 pKa = 4.21LTDD685 pKa = 3.4VRR687 pKa = 11.84APEE690 pKa = 4.31LPRR693 pKa = 11.84ALANSGVSVSPSAVQAAKK711 pKa = 10.16DD712 pKa = 3.79GKK714 pKa = 9.2MSAIKK719 pKa = 10.46VIAAASALARR729 pKa = 11.84ASDD732 pKa = 3.83FSGILPTVSNKK743 pKa = 9.53YY744 pKa = 9.31LQYY747 pKa = 11.45ANEE750 pKa = 4.35CSKK753 pKa = 11.18SDD755 pKa = 3.71YY756 pKa = 10.85VDD758 pKa = 3.73RR759 pKa = 11.84EE760 pKa = 3.81CSIRR764 pKa = 11.84CFGFDD769 pKa = 2.94GHH771 pKa = 5.79SAKK774 pKa = 10.05EE775 pKa = 3.68VRR777 pKa = 11.84EE778 pKa = 4.15RR779 pKa = 11.84IEE781 pKa = 3.78EE782 pKa = 4.07RR783 pKa = 11.84NLGVTHH789 pKa = 6.82QDD791 pKa = 3.13EE792 pKa = 4.64QKK794 pKa = 8.66TLEE797 pKa = 4.02MLGFKK802 pKa = 9.06ATHH805 pKa = 6.97DD806 pKa = 3.58EE807 pKa = 4.4LMTFTEE813 pKa = 5.29HH814 pKa = 7.16IWDD817 pKa = 4.77LEE819 pKa = 4.09PATLTAYY826 pKa = 10.05QAFRR830 pKa = 11.84EE831 pKa = 4.25SLPASWRR838 pKa = 11.84MEE840 pKa = 3.85
MM1 pKa = 7.36LSVRR5 pKa = 11.84EE6 pKa = 4.23GFSDD10 pKa = 3.92LKK12 pKa = 10.33YY13 pKa = 11.25GEE15 pKa = 4.17VAPYY19 pKa = 10.24LRR21 pKa = 11.84EE22 pKa = 3.83RR23 pKa = 11.84AAKK26 pKa = 9.99RR27 pKa = 11.84QGEE30 pKa = 4.27WTLTVHH36 pKa = 6.0EE37 pKa = 5.67AEE39 pKa = 4.93DD40 pKa = 3.44SWLGILSRR48 pKa = 11.84AVSFKK53 pKa = 10.98VVLTEE58 pKa = 3.73EE59 pKa = 4.88EE60 pKa = 4.31YY61 pKa = 11.4AKK63 pKa = 10.67LSRR66 pKa = 11.84ALYY69 pKa = 9.61HH70 pKa = 7.03GIRR73 pKa = 11.84NGTKK77 pKa = 8.8KK78 pKa = 9.31TSLLANLEE86 pKa = 4.03NTAQALWPTGSLDD99 pKa = 3.55HH100 pKa = 6.81VAMRR104 pKa = 11.84HH105 pKa = 4.8LAVTAVVQYY114 pKa = 7.36VTKK117 pKa = 10.44EE118 pKa = 3.77STPQGFRR125 pKa = 11.84LPYY128 pKa = 8.31WMTWGAMVTQATVGVHH144 pKa = 4.45ILGRR148 pKa = 11.84LVKK151 pKa = 10.35GAARR155 pKa = 11.84RR156 pKa = 11.84AWITGPHH163 pKa = 6.99PGPFGQLIMRR173 pKa = 11.84EE174 pKa = 4.28YY175 pKa = 10.32FATPALVMWALMTVISYY192 pKa = 10.52CGARR196 pKa = 11.84GITLSWRR203 pKa = 11.84SVLPVLSRR211 pKa = 11.84NAGTTRR217 pKa = 11.84TGPAAEE223 pKa = 4.72AAPAAPGGPPPAGATPGAGGAAPGPAGGAPPSGGTPAAGAPAPPSPPVMEE273 pKa = 4.43QPATPEE279 pKa = 4.12DD280 pKa = 4.01PEE282 pKa = 4.09NAANASEE289 pKa = 4.39DD290 pKa = 3.81LRR292 pKa = 11.84TMEE295 pKa = 4.38MPGKK299 pKa = 9.9IVGVLGQNFDD309 pKa = 3.24KK310 pKa = 10.92DD311 pKa = 3.91LPVNHH316 pKa = 6.78LPVVGVLVGPCQRR329 pKa = 11.84KK330 pKa = 8.28PNVYY334 pKa = 10.3SKK336 pKa = 9.71TVEE339 pKa = 4.0NLKK342 pKa = 10.56AAINEE347 pKa = 4.26RR348 pKa = 11.84IIKK351 pKa = 9.84KK352 pKa = 9.6ARR354 pKa = 11.84KK355 pKa = 8.64PKK357 pKa = 9.95LCRR360 pKa = 11.84ADD362 pKa = 3.37RR363 pKa = 11.84VRR365 pKa = 11.84ISRR368 pKa = 11.84LVRR371 pKa = 11.84KK372 pKa = 9.96AMSNSEE378 pKa = 3.84KK379 pKa = 10.61HH380 pKa = 6.06GVFSKK385 pKa = 10.94SRR387 pKa = 11.84IQEE390 pKa = 3.46WAIQHH395 pKa = 5.96FNLEE399 pKa = 4.26EE400 pKa = 4.01MKK402 pKa = 10.26SQKK405 pKa = 9.45WSLEE409 pKa = 3.81RR410 pKa = 11.84FRR412 pKa = 11.84NALKK416 pKa = 10.58NLYY419 pKa = 9.78EE420 pKa = 4.65KK421 pKa = 11.14DD422 pKa = 3.47PVEE425 pKa = 4.51FMLKK429 pKa = 10.47AGIKK433 pKa = 8.23PEE435 pKa = 3.94CMEE438 pKa = 4.21EE439 pKa = 4.29GKK441 pKa = 10.61APRR444 pKa = 11.84MLIADD449 pKa = 3.91GDD451 pKa = 4.11EE452 pKa = 4.41GQLMALAVIKK462 pKa = 10.61CFEE465 pKa = 4.11EE466 pKa = 4.39LLFKK470 pKa = 10.82HH471 pKa = 6.16FEE473 pKa = 3.99EE474 pKa = 5.51RR475 pKa = 11.84SIKK478 pKa = 10.33HH479 pKa = 5.45LSKK482 pKa = 10.65HH483 pKa = 4.81EE484 pKa = 3.85AMEE487 pKa = 4.0RR488 pKa = 11.84VVRR491 pKa = 11.84VLKK494 pKa = 10.84KK495 pKa = 9.43KK496 pKa = 9.74GARR499 pKa = 11.84AVEE502 pKa = 4.36GDD504 pKa = 3.23GSAWDD509 pKa = 3.99TTCGVLVRR517 pKa = 11.84SLIEE521 pKa = 3.88NPILFHH527 pKa = 7.03IFRR530 pKa = 11.84TLAEE534 pKa = 4.3FGVIPEE540 pKa = 3.86TWMAEE545 pKa = 3.64HH546 pKa = 6.86HH547 pKa = 6.15AACTKK552 pKa = 10.58EE553 pKa = 3.72KK554 pKa = 10.95LKK556 pKa = 11.22LFFSNKK562 pKa = 6.27FQKK565 pKa = 9.2VTVTIDD571 pKa = 3.78AIRR574 pKa = 11.84RR575 pKa = 11.84SGHH578 pKa = 6.55RR579 pKa = 11.84GTSCLNWWINFTMWVSSIFKK599 pKa = 9.87EE600 pKa = 4.25PEE602 pKa = 3.66RR603 pKa = 11.84FLDD606 pKa = 3.49PDD608 pKa = 3.5VRR610 pKa = 11.84KK611 pKa = 8.86GTDD614 pKa = 3.13LTEE617 pKa = 4.62KK618 pKa = 9.66EE619 pKa = 4.24RR620 pKa = 11.84WWNGTFEE627 pKa = 5.69GDD629 pKa = 4.45DD630 pKa = 4.39SLCTMCPPMLKK641 pKa = 9.7DD642 pKa = 4.93DD643 pKa = 3.87KK644 pKa = 10.92MSDD647 pKa = 3.23IFCQFWSDD655 pKa = 2.97AGFNMKK661 pKa = 9.61IVYY664 pKa = 9.59CDD666 pKa = 3.13RR667 pKa = 11.84RR668 pKa = 11.84ATFTGWHH675 pKa = 5.94LVCEE679 pKa = 4.38NGEE682 pKa = 4.21LTDD685 pKa = 3.4VRR687 pKa = 11.84APEE690 pKa = 4.31LPRR693 pKa = 11.84ALANSGVSVSPSAVQAAKK711 pKa = 10.16DD712 pKa = 3.79GKK714 pKa = 9.2MSAIKK719 pKa = 10.46VIAAASALARR729 pKa = 11.84ASDD732 pKa = 3.83FSGILPTVSNKK743 pKa = 9.53YY744 pKa = 9.31LQYY747 pKa = 11.45ANEE750 pKa = 4.35CSKK753 pKa = 11.18SDD755 pKa = 3.71YY756 pKa = 10.85VDD758 pKa = 3.73RR759 pKa = 11.84EE760 pKa = 3.81CSIRR764 pKa = 11.84CFGFDD769 pKa = 2.94GHH771 pKa = 5.79SAKK774 pKa = 10.05EE775 pKa = 3.68VRR777 pKa = 11.84EE778 pKa = 4.15RR779 pKa = 11.84IEE781 pKa = 3.78EE782 pKa = 4.07RR783 pKa = 11.84NLGVTHH789 pKa = 6.82QDD791 pKa = 3.13EE792 pKa = 4.64QKK794 pKa = 8.66TLEE797 pKa = 4.02MLGFKK802 pKa = 9.06ATHH805 pKa = 6.97DD806 pKa = 3.58EE807 pKa = 4.4LMTFTEE813 pKa = 5.29HH814 pKa = 7.16IWDD817 pKa = 4.77LEE819 pKa = 4.09PATLTAYY826 pKa = 10.05QAFRR830 pKa = 11.84EE831 pKa = 4.25SLPASWRR838 pKa = 11.84MEE840 pKa = 3.85
Molecular weight: 93.25 kDa
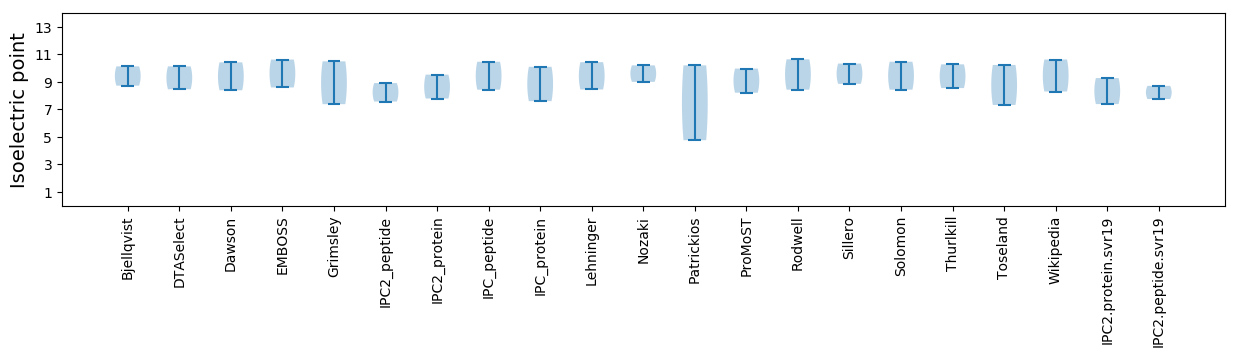
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL43|A0A1L3KL43_9VIRU RNA-dependent RNA polymerase OS=Wenzhou weivirus-like virus 1 OX=1923682 PE=4 SV=1
MM1 pKa = 7.41ARR3 pKa = 11.84GGRR6 pKa = 11.84RR7 pKa = 11.84AGRR10 pKa = 11.84AKK12 pKa = 10.42AQPKK16 pKa = 9.39PEE18 pKa = 4.19KK19 pKa = 9.8KK20 pKa = 10.32AQGGQTSAPRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 9.64RR33 pKa = 11.84RR34 pKa = 11.84GRR36 pKa = 11.84PNSGWAVNGRR46 pKa = 11.84AHH48 pKa = 6.81HH49 pKa = 5.45GHH51 pKa = 6.45YY52 pKa = 10.62DD53 pKa = 3.15AFDD56 pKa = 3.77PRR58 pKa = 11.84KK59 pKa = 9.78QPMARR64 pKa = 11.84AVGHH68 pKa = 5.43ATAVRR73 pKa = 11.84GFGRR77 pKa = 11.84LKK79 pKa = 10.49RR80 pKa = 11.84PSFNDD85 pKa = 2.79GMEE88 pKa = 3.9QMYY91 pKa = 10.7VFMPSTTRR99 pKa = 11.84VVGLYY104 pKa = 10.81GNYY107 pKa = 10.04KK108 pKa = 10.32SDD110 pKa = 3.07IDD112 pKa = 3.49QWRR115 pKa = 11.84GAGPNVDD122 pKa = 3.56TNTALIEE129 pKa = 3.98NMGFSEE135 pKa = 4.86EE136 pKa = 4.4SGPHH140 pKa = 3.81QTLLGKK146 pKa = 10.32FSVRR150 pKa = 11.84LRR152 pKa = 11.84NISKK156 pKa = 10.8AMDD159 pKa = 3.64ASGSVYY165 pKa = 10.66VLSLNAGMFLEE176 pKa = 4.63SLNGDD181 pKa = 2.99WTARR185 pKa = 11.84TNWTRR190 pKa = 11.84LRR192 pKa = 11.84NYY194 pKa = 10.19VLGSPRR200 pKa = 11.84TRR202 pKa = 11.84TFSGSEE208 pKa = 3.87LLKK211 pKa = 8.78TRR213 pKa = 11.84QWNTHH218 pKa = 6.75PIDD221 pKa = 3.49ATRR224 pKa = 11.84SQEE227 pKa = 4.04FQMVPPATPDD237 pKa = 3.41TQVTTFRR244 pKa = 11.84NSMDD248 pKa = 3.81SPTHH252 pKa = 6.51SMVVFLFIPGEE263 pKa = 4.31SNVRR267 pKa = 11.84NMYY270 pKa = 9.89EE271 pKa = 3.61FSFAANHH278 pKa = 4.94YY279 pKa = 9.87CRR281 pKa = 11.84YY282 pKa = 9.22EE283 pKa = 4.17VNGPLANAAEE293 pKa = 4.54PVPTAPISVIDD304 pKa = 3.87GARR307 pKa = 11.84KK308 pKa = 8.68FVEE311 pKa = 4.24NVGSLGHH318 pKa = 5.41VVEE321 pKa = 5.15DD322 pKa = 3.71VVGTINAYY330 pKa = 10.24APAGLL335 pKa = 4.01
MM1 pKa = 7.41ARR3 pKa = 11.84GGRR6 pKa = 11.84RR7 pKa = 11.84AGRR10 pKa = 11.84AKK12 pKa = 10.42AQPKK16 pKa = 9.39PEE18 pKa = 4.19KK19 pKa = 9.8KK20 pKa = 10.32AQGGQTSAPRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 9.64RR33 pKa = 11.84RR34 pKa = 11.84GRR36 pKa = 11.84PNSGWAVNGRR46 pKa = 11.84AHH48 pKa = 6.81HH49 pKa = 5.45GHH51 pKa = 6.45YY52 pKa = 10.62DD53 pKa = 3.15AFDD56 pKa = 3.77PRR58 pKa = 11.84KK59 pKa = 9.78QPMARR64 pKa = 11.84AVGHH68 pKa = 5.43ATAVRR73 pKa = 11.84GFGRR77 pKa = 11.84LKK79 pKa = 10.49RR80 pKa = 11.84PSFNDD85 pKa = 2.79GMEE88 pKa = 3.9QMYY91 pKa = 10.7VFMPSTTRR99 pKa = 11.84VVGLYY104 pKa = 10.81GNYY107 pKa = 10.04KK108 pKa = 10.32SDD110 pKa = 3.07IDD112 pKa = 3.49QWRR115 pKa = 11.84GAGPNVDD122 pKa = 3.56TNTALIEE129 pKa = 3.98NMGFSEE135 pKa = 4.86EE136 pKa = 4.4SGPHH140 pKa = 3.81QTLLGKK146 pKa = 10.32FSVRR150 pKa = 11.84LRR152 pKa = 11.84NISKK156 pKa = 10.8AMDD159 pKa = 3.64ASGSVYY165 pKa = 10.66VLSLNAGMFLEE176 pKa = 4.63SLNGDD181 pKa = 2.99WTARR185 pKa = 11.84TNWTRR190 pKa = 11.84LRR192 pKa = 11.84NYY194 pKa = 10.19VLGSPRR200 pKa = 11.84TRR202 pKa = 11.84TFSGSEE208 pKa = 3.87LLKK211 pKa = 8.78TRR213 pKa = 11.84QWNTHH218 pKa = 6.75PIDD221 pKa = 3.49ATRR224 pKa = 11.84SQEE227 pKa = 4.04FQMVPPATPDD237 pKa = 3.41TQVTTFRR244 pKa = 11.84NSMDD248 pKa = 3.81SPTHH252 pKa = 6.51SMVVFLFIPGEE263 pKa = 4.31SNVRR267 pKa = 11.84NMYY270 pKa = 9.89EE271 pKa = 3.61FSFAANHH278 pKa = 4.94YY279 pKa = 9.87CRR281 pKa = 11.84YY282 pKa = 9.22EE283 pKa = 4.17VNGPLANAAEE293 pKa = 4.54PVPTAPISVIDD304 pKa = 3.87GARR307 pKa = 11.84KK308 pKa = 8.68FVEE311 pKa = 4.24NVGSLGHH318 pKa = 5.41VVEE321 pKa = 5.15DD322 pKa = 3.71VVGTINAYY330 pKa = 10.24APAGLL335 pKa = 4.01
Molecular weight: 36.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1175 |
335 |
840 |
587.5 |
65.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.043 ± 0.374 | 1.447 ± 0.545 |
3.915 ± 0.016 | 6.638 ± 1.166 |
4.085 ± 0.186 | 7.915 ± 0.776 |
2.553 ± 0.063 | 3.574 ± 0.563 |
5.532 ± 0.925 | 7.404 ± 0.963 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.319 ± 0.125 | 4.0 ± 1.076 |
6.128 ± 0.208 | 2.638 ± 0.306 |
7.149 ± 0.857 | 6.383 ± 0.37 |
6.043 ± 0.249 | 6.894 ± 0.27 |
2.213 ± 0.342 | 2.128 ± 0.407 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
