
Ferroglobus placidus (strain DSM 10642 / AEDII12DO)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Archaeoglobi; Archaeoglobales; Archaeoglobaceae; Ferroglobus; Ferroglobus placidus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
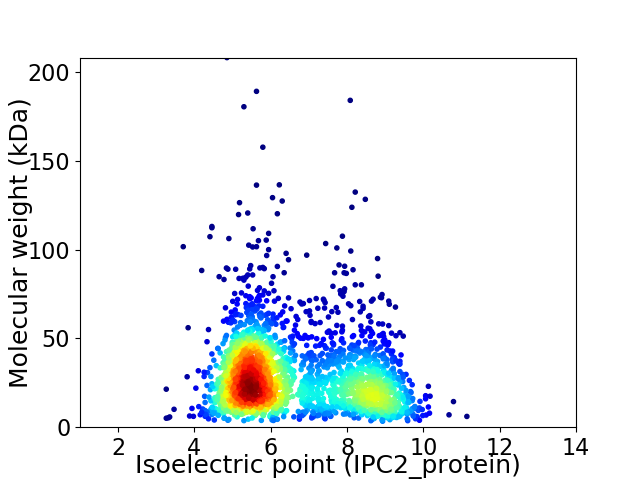
Virtual 2D-PAGE plot for 2463 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D3RXM1|D3RXM1_FERPA Chemotaxis protein CheA OS=Ferroglobus placidus (strain DSM 10642 / AEDII12DO) OX=589924 GN=Ferp_1075 PE=4 SV=1
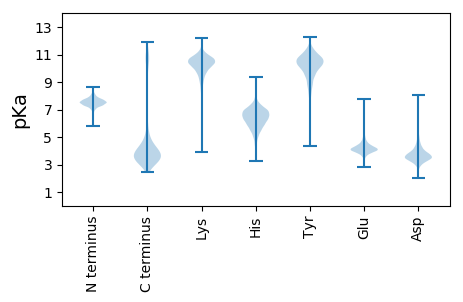
MM1 pKa = 7.14GKK3 pKa = 8.85VWKK6 pKa = 10.46VGLAVLVTLLITMATANAIQPSSIFNNSSLTVTIEE41 pKa = 3.96PNPTFAGTPIVSPVNFSVVNAGWYY65 pKa = 10.46GDD67 pKa = 3.49IPITFNVTSPNMGNVTVNVSTDD89 pKa = 3.14LTAYY93 pKa = 10.33NSSTSGTSIMLNLTTEE109 pKa = 3.54TWFAVNASTAGCYY122 pKa = 7.28WVNVSHH128 pKa = 7.79DD129 pKa = 3.71LTNDD133 pKa = 3.2YY134 pKa = 11.69VNFTVCYY141 pKa = 8.41WKK143 pKa = 10.03IDD145 pKa = 3.63SVTLYY150 pKa = 10.62DD151 pKa = 4.17VNGNSYY157 pKa = 11.3DD158 pKa = 3.76LLAGGTQNIEE168 pKa = 3.77AGDD171 pKa = 3.6YY172 pKa = 9.66TIYY175 pKa = 10.78INVTAPSGEE184 pKa = 4.2GYY186 pKa = 10.23EE187 pKa = 4.25VANASLNLTPFGIGWVTLDD206 pKa = 4.44VIDD209 pKa = 4.63SNSTAGTYY217 pKa = 10.18NLSGTIQFHH226 pKa = 6.59KK227 pKa = 10.53ATDD230 pKa = 3.17AGYY233 pKa = 10.6NITGNITIGDD243 pKa = 4.06PVEE246 pKa = 4.45IPAINTSQVDD256 pKa = 3.75VDD258 pKa = 3.83PAPAVKK264 pKa = 9.18WAFLEE269 pKa = 4.3TNPYY273 pKa = 10.92DD274 pKa = 4.53SDD276 pKa = 5.81DD277 pKa = 3.73NVIWDD282 pKa = 4.06GVSDD286 pKa = 4.3YY287 pKa = 11.51INATLYY293 pKa = 10.1VAAVDD298 pKa = 3.59EE299 pKa = 4.77FGNINTSGFSGYY311 pKa = 9.92ISVVSGPGILNKK323 pKa = 10.14TSVNASRR330 pKa = 11.84VDD332 pKa = 3.23AFNISVPNADD342 pKa = 2.65ATAFVTVKK350 pKa = 10.61VYY352 pKa = 11.01GGGLASTTKK361 pKa = 10.2TVGFYY366 pKa = 11.15DD367 pKa = 5.28RR368 pKa = 11.84IDD370 pKa = 3.87HH371 pKa = 7.21LDD373 pKa = 3.48VAVEE377 pKa = 4.12KK378 pKa = 10.54PVLYY382 pKa = 10.79ANNSDD387 pKa = 3.58STTVYY392 pKa = 9.2VTLKK396 pKa = 10.66DD397 pKa = 3.59SAGNTIPVEE406 pKa = 4.48GINITIDD413 pKa = 3.17EE414 pKa = 4.66LTTLGLDD421 pKa = 3.37IPTSTNATDD430 pKa = 3.51NSGVATFTVTAGANSGTATIGAIVDD455 pKa = 4.32SGIAWDD461 pKa = 4.5GRR463 pKa = 11.84YY464 pKa = 10.28GDD466 pKa = 4.92ANITLKK472 pKa = 10.44QAPDD476 pKa = 3.4TTAVQTAATSAVPSTITAGEE496 pKa = 4.57GYY498 pKa = 10.0KK499 pKa = 9.78IEE501 pKa = 4.23FTIVDD506 pKa = 3.67HH507 pKa = 6.99AGQPIANSAEE517 pKa = 4.01FPVKK521 pKa = 10.45FNITAGDD528 pKa = 4.04AVWLEE533 pKa = 4.05NNAKK537 pKa = 10.11VLFVNATDD545 pKa = 3.86ANGNVSATLYY555 pKa = 8.81STNASTSINVNISIKK570 pKa = 10.54NEE572 pKa = 3.59SGQWVTLVPDD582 pKa = 3.79TPVNVDD588 pKa = 3.44PAEE591 pKa = 4.12VADD594 pKa = 5.11FVVEE598 pKa = 3.99PDD600 pKa = 4.11SIGLPAVIGASEE612 pKa = 4.57VINVTLVDD620 pKa = 3.84AYY622 pKa = 11.08GNVNSTNATMIVTTDD637 pKa = 3.44NPSLGLLNNGTNHH650 pKa = 7.28PDD652 pKa = 3.61SIVVSITGGKK662 pKa = 10.45GSFTYY667 pKa = 10.34YY668 pKa = 10.65VNSSTPGVANLTLNVTDD685 pKa = 3.93FNIVKK690 pKa = 8.55TVKK693 pKa = 10.08IVTTAPQGVVLTFDD707 pKa = 3.04QTLPFVASDD716 pKa = 3.48INVTAQVTTAEE727 pKa = 4.3GLPLKK732 pKa = 10.4YY733 pKa = 10.74SNVDD737 pKa = 3.14LTLVVKK743 pKa = 10.9NPDD746 pKa = 3.19GNVIFIGTASTNASGAAVWTLNGTNFTAPGVYY778 pKa = 8.81TFKK781 pKa = 10.99VYY783 pKa = 8.83NTSYY787 pKa = 10.81GISDD791 pKa = 3.88TKK793 pKa = 10.97QLTFAGPAVKK803 pKa = 10.47VVVTVNNTAPLVNDD817 pKa = 3.63TVMITATFYY826 pKa = 11.22DD827 pKa = 3.96VNDD830 pKa = 4.42LVTSDD835 pKa = 5.12LDD837 pKa = 3.93GADD840 pKa = 3.34VTFLINDD847 pKa = 3.79VVVGTATITNGVASINYY864 pKa = 7.22TFTEE868 pKa = 4.21AGTFNITAFYY878 pKa = 10.77NATLQDD884 pKa = 4.1TIAITVVTTVAPGDD898 pKa = 3.84SDD900 pKa = 4.08NDD902 pKa = 3.33GWNDD906 pKa = 3.39SVEE909 pKa = 4.1ALIASYY915 pKa = 11.04NPSFDD920 pKa = 3.71PAQVPTQDD928 pKa = 3.79DD929 pKa = 4.13VINAVVGAVNKK940 pKa = 10.38YY941 pKa = 9.76FDD943 pKa = 4.11PATTEE948 pKa = 3.99SEE950 pKa = 4.01KK951 pKa = 10.58QQIVNDD957 pKa = 4.59IITLVQEE964 pKa = 4.41YY965 pKa = 9.93FLHH968 pKa = 6.69FF969 pKa = 4.1
MM1 pKa = 7.14GKK3 pKa = 8.85VWKK6 pKa = 10.46VGLAVLVTLLITMATANAIQPSSIFNNSSLTVTIEE41 pKa = 3.96PNPTFAGTPIVSPVNFSVVNAGWYY65 pKa = 10.46GDD67 pKa = 3.49IPITFNVTSPNMGNVTVNVSTDD89 pKa = 3.14LTAYY93 pKa = 10.33NSSTSGTSIMLNLTTEE109 pKa = 3.54TWFAVNASTAGCYY122 pKa = 7.28WVNVSHH128 pKa = 7.79DD129 pKa = 3.71LTNDD133 pKa = 3.2YY134 pKa = 11.69VNFTVCYY141 pKa = 8.41WKK143 pKa = 10.03IDD145 pKa = 3.63SVTLYY150 pKa = 10.62DD151 pKa = 4.17VNGNSYY157 pKa = 11.3DD158 pKa = 3.76LLAGGTQNIEE168 pKa = 3.77AGDD171 pKa = 3.6YY172 pKa = 9.66TIYY175 pKa = 10.78INVTAPSGEE184 pKa = 4.2GYY186 pKa = 10.23EE187 pKa = 4.25VANASLNLTPFGIGWVTLDD206 pKa = 4.44VIDD209 pKa = 4.63SNSTAGTYY217 pKa = 10.18NLSGTIQFHH226 pKa = 6.59KK227 pKa = 10.53ATDD230 pKa = 3.17AGYY233 pKa = 10.6NITGNITIGDD243 pKa = 4.06PVEE246 pKa = 4.45IPAINTSQVDD256 pKa = 3.75VDD258 pKa = 3.83PAPAVKK264 pKa = 9.18WAFLEE269 pKa = 4.3TNPYY273 pKa = 10.92DD274 pKa = 4.53SDD276 pKa = 5.81DD277 pKa = 3.73NVIWDD282 pKa = 4.06GVSDD286 pKa = 4.3YY287 pKa = 11.51INATLYY293 pKa = 10.1VAAVDD298 pKa = 3.59EE299 pKa = 4.77FGNINTSGFSGYY311 pKa = 9.92ISVVSGPGILNKK323 pKa = 10.14TSVNASRR330 pKa = 11.84VDD332 pKa = 3.23AFNISVPNADD342 pKa = 2.65ATAFVTVKK350 pKa = 10.61VYY352 pKa = 11.01GGGLASTTKK361 pKa = 10.2TVGFYY366 pKa = 11.15DD367 pKa = 5.28RR368 pKa = 11.84IDD370 pKa = 3.87HH371 pKa = 7.21LDD373 pKa = 3.48VAVEE377 pKa = 4.12KK378 pKa = 10.54PVLYY382 pKa = 10.79ANNSDD387 pKa = 3.58STTVYY392 pKa = 9.2VTLKK396 pKa = 10.66DD397 pKa = 3.59SAGNTIPVEE406 pKa = 4.48GINITIDD413 pKa = 3.17EE414 pKa = 4.66LTTLGLDD421 pKa = 3.37IPTSTNATDD430 pKa = 3.51NSGVATFTVTAGANSGTATIGAIVDD455 pKa = 4.32SGIAWDD461 pKa = 4.5GRR463 pKa = 11.84YY464 pKa = 10.28GDD466 pKa = 4.92ANITLKK472 pKa = 10.44QAPDD476 pKa = 3.4TTAVQTAATSAVPSTITAGEE496 pKa = 4.57GYY498 pKa = 10.0KK499 pKa = 9.78IEE501 pKa = 4.23FTIVDD506 pKa = 3.67HH507 pKa = 6.99AGQPIANSAEE517 pKa = 4.01FPVKK521 pKa = 10.45FNITAGDD528 pKa = 4.04AVWLEE533 pKa = 4.05NNAKK537 pKa = 10.11VLFVNATDD545 pKa = 3.86ANGNVSATLYY555 pKa = 8.81STNASTSINVNISIKK570 pKa = 10.54NEE572 pKa = 3.59SGQWVTLVPDD582 pKa = 3.79TPVNVDD588 pKa = 3.44PAEE591 pKa = 4.12VADD594 pKa = 5.11FVVEE598 pKa = 3.99PDD600 pKa = 4.11SIGLPAVIGASEE612 pKa = 4.57VINVTLVDD620 pKa = 3.84AYY622 pKa = 11.08GNVNSTNATMIVTTDD637 pKa = 3.44NPSLGLLNNGTNHH650 pKa = 7.28PDD652 pKa = 3.61SIVVSITGGKK662 pKa = 10.45GSFTYY667 pKa = 10.34YY668 pKa = 10.65VNSSTPGVANLTLNVTDD685 pKa = 3.93FNIVKK690 pKa = 8.55TVKK693 pKa = 10.08IVTTAPQGVVLTFDD707 pKa = 3.04QTLPFVASDD716 pKa = 3.48INVTAQVTTAEE727 pKa = 4.3GLPLKK732 pKa = 10.4YY733 pKa = 10.74SNVDD737 pKa = 3.14LTLVVKK743 pKa = 10.9NPDD746 pKa = 3.19GNVIFIGTASTNASGAAVWTLNGTNFTAPGVYY778 pKa = 8.81TFKK781 pKa = 10.99VYY783 pKa = 8.83NTSYY787 pKa = 10.81GISDD791 pKa = 3.88TKK793 pKa = 10.97QLTFAGPAVKK803 pKa = 10.47VVVTVNNTAPLVNDD817 pKa = 3.63TVMITATFYY826 pKa = 11.22DD827 pKa = 3.96VNDD830 pKa = 4.42LVTSDD835 pKa = 5.12LDD837 pKa = 3.93GADD840 pKa = 3.34VTFLINDD847 pKa = 3.79VVVGTATITNGVASINYY864 pKa = 7.22TFTEE868 pKa = 4.21AGTFNITAFYY878 pKa = 10.77NATLQDD884 pKa = 4.1TIAITVVTTVAPGDD898 pKa = 3.84SDD900 pKa = 4.08NDD902 pKa = 3.33GWNDD906 pKa = 3.39SVEE909 pKa = 4.1ALIASYY915 pKa = 11.04NPSFDD920 pKa = 3.71PAQVPTQDD928 pKa = 3.79DD929 pKa = 4.13VINAVVGAVNKK940 pKa = 10.38YY941 pKa = 9.76FDD943 pKa = 4.11PATTEE948 pKa = 3.99SEE950 pKa = 4.01KK951 pKa = 10.58QQIVNDD957 pKa = 4.59IITLVQEE964 pKa = 4.41YY965 pKa = 9.93FLHH968 pKa = 6.69FF969 pKa = 4.1
Molecular weight: 101.74 kDa
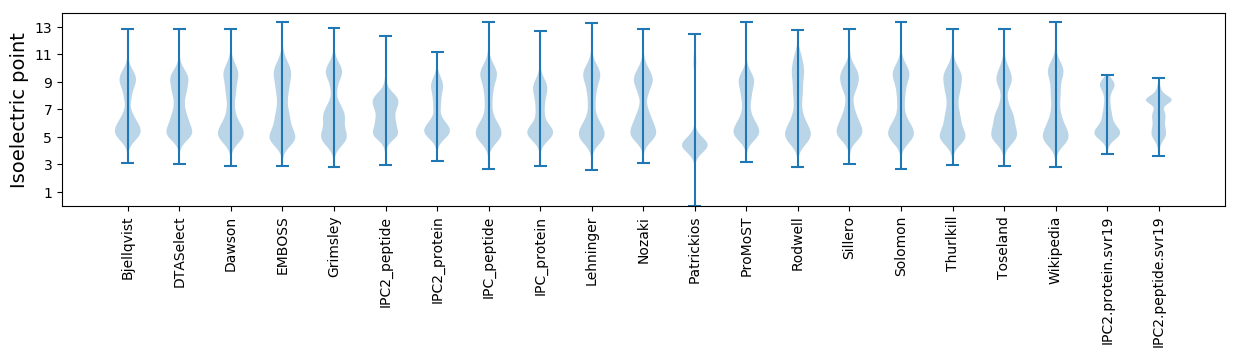
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D3S0Z0|D3S0Z0_FERPA CRM domain-containing protein OS=Ferroglobus placidus (strain DSM 10642 / AEDII12DO) OX=589924 GN=Ferp_0034 PE=4 SV=1
MM1 pKa = 7.34GKK3 pKa = 8.36KK4 pKa = 8.33TVGVKK9 pKa = 10.11KK10 pKa = 10.67RR11 pKa = 11.84LGKK14 pKa = 9.83FLKK17 pKa = 10.06QNRR20 pKa = 11.84RR21 pKa = 11.84PPVWVILKK29 pKa = 6.93TKK31 pKa = 10.0RR32 pKa = 11.84RR33 pKa = 11.84VVTSPKK39 pKa = 9.41RR40 pKa = 11.84RR41 pKa = 11.84FWRR44 pKa = 11.84HH45 pKa = 4.28TKK47 pKa = 10.74LKK49 pKa = 10.26VV50 pKa = 2.91
MM1 pKa = 7.34GKK3 pKa = 8.36KK4 pKa = 8.33TVGVKK9 pKa = 10.11KK10 pKa = 10.67RR11 pKa = 11.84LGKK14 pKa = 9.83FLKK17 pKa = 10.06QNRR20 pKa = 11.84RR21 pKa = 11.84PPVWVILKK29 pKa = 6.93TKK31 pKa = 10.0RR32 pKa = 11.84RR33 pKa = 11.84VVTSPKK39 pKa = 9.41RR40 pKa = 11.84RR41 pKa = 11.84FWRR44 pKa = 11.84HH45 pKa = 4.28TKK47 pKa = 10.74LKK49 pKa = 10.26VV50 pKa = 2.91
Molecular weight: 6.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
660597 |
31 |
1864 |
268.2 |
30.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.075 ± 0.053 | 1.128 ± 0.028 |
4.758 ± 0.033 | 9.528 ± 0.081 |
4.764 ± 0.045 | 6.73 ± 0.04 |
1.487 ± 0.021 | 7.697 ± 0.049 |
8.438 ± 0.056 | 9.446 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.2 ± 0.022 | 3.387 ± 0.033 |
3.687 ± 0.028 | 1.5 ± 0.02 |
5.252 ± 0.048 | 5.525 ± 0.048 |
3.965 ± 0.043 | 8.579 ± 0.044 |
1.035 ± 0.024 | 3.821 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
