
Bovine faeces associated circular DNA virus 2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
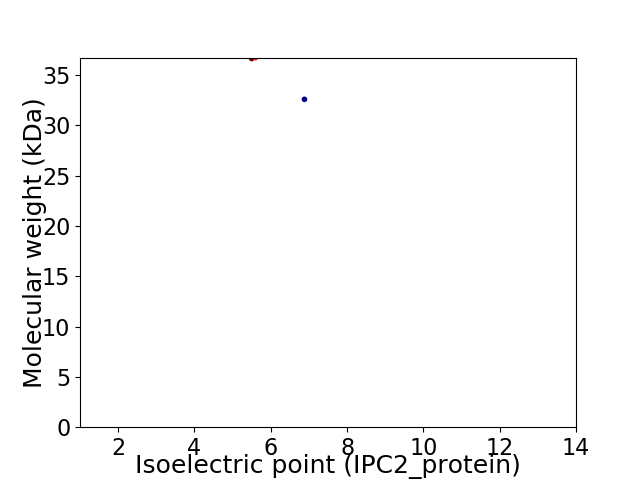
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MFX8|A0A168MFX8_9VIRU Capsid protein OS=Bovine faeces associated circular DNA virus 2 OX=1843765 PE=4 SV=1
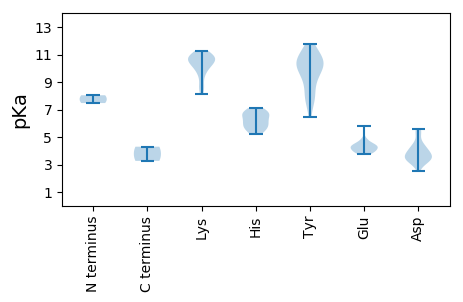
MM1 pKa = 7.47SVRR4 pKa = 11.84DD5 pKa = 3.97YY6 pKa = 11.16RR7 pKa = 11.84SKK9 pKa = 11.28NYY11 pKa = 10.27LLTYY15 pKa = 8.41PRR17 pKa = 11.84CGLQQWWVLTNLQNNEE33 pKa = 4.37RR34 pKa = 11.84IKK36 pKa = 10.5MIIQYY41 pKa = 9.97VIVAQEE47 pKa = 3.81NHH49 pKa = 5.65QDD51 pKa = 3.7GAEE54 pKa = 4.43HH55 pKa = 5.95IHH57 pKa = 6.25VFLALTEE64 pKa = 4.54PIRR67 pKa = 11.84IKK69 pKa = 10.84SNEE72 pKa = 3.81MDD74 pKa = 3.25VFDD77 pKa = 5.62IVDD80 pKa = 3.41EE81 pKa = 4.58DD82 pKa = 4.19EE83 pKa = 4.38PDD85 pKa = 3.2KK86 pKa = 11.01RR87 pKa = 11.84VYY89 pKa = 10.09HH90 pKa = 6.55CNIQSVKK97 pKa = 10.29SPKK100 pKa = 10.16DD101 pKa = 3.35AIQYY105 pKa = 7.82VKK107 pKa = 10.94KK108 pKa = 9.7NGHH111 pKa = 6.84FITYY115 pKa = 7.76GICPWKK121 pKa = 10.87VPLTMAEE128 pKa = 4.31KK129 pKa = 11.09NEE131 pKa = 3.88LLQSKK136 pKa = 9.54SLHH139 pKa = 5.75EE140 pKa = 4.09LVEE143 pKa = 4.37SGMVSIFKK151 pKa = 10.32IPQLQKK157 pKa = 10.86AISILQNEE165 pKa = 4.82LGNVEE170 pKa = 4.02RR171 pKa = 11.84QPPKK175 pKa = 8.47VHH177 pKa = 6.78WYY179 pKa = 9.64YY180 pKa = 11.5GSTGTGKK187 pKa = 8.11TRR189 pKa = 11.84TAYY192 pKa = 10.07EE193 pKa = 3.94EE194 pKa = 4.24AKK196 pKa = 10.46EE197 pKa = 4.33SEE199 pKa = 3.88DD200 pKa = 4.42GYY202 pKa = 10.34WISHH206 pKa = 5.23EE207 pKa = 4.1TGEE210 pKa = 4.47WFDD213 pKa = 4.39GYY215 pKa = 9.91IGQGIAILDD224 pKa = 4.7DD225 pKa = 3.05IRR227 pKa = 11.84AATWPLQKK235 pKa = 10.63LLRR238 pKa = 11.84LCDD241 pKa = 3.95RR242 pKa = 11.84YY243 pKa = 10.25PIQVPIKK250 pKa = 10.79GGFTWWRR257 pKa = 11.84PRR259 pKa = 11.84EE260 pKa = 4.03IYY262 pKa = 8.78ITAPGRR268 pKa = 11.84PEE270 pKa = 5.81DD271 pKa = 3.3IYY273 pKa = 11.75KK274 pKa = 10.68NYY276 pKa = 7.75TTGQPYY282 pKa = 11.2DD283 pKa = 3.97NIEE286 pKa = 3.86QLNRR290 pKa = 11.84RR291 pKa = 11.84ITEE294 pKa = 4.14MIDD297 pKa = 3.26FDD299 pKa = 5.54SEE301 pKa = 4.51TEE303 pKa = 4.74PIDD306 pKa = 3.39QTLYY310 pKa = 10.21WKK312 pKa = 10.71GG313 pKa = 3.26
MM1 pKa = 7.47SVRR4 pKa = 11.84DD5 pKa = 3.97YY6 pKa = 11.16RR7 pKa = 11.84SKK9 pKa = 11.28NYY11 pKa = 10.27LLTYY15 pKa = 8.41PRR17 pKa = 11.84CGLQQWWVLTNLQNNEE33 pKa = 4.37RR34 pKa = 11.84IKK36 pKa = 10.5MIIQYY41 pKa = 9.97VIVAQEE47 pKa = 3.81NHH49 pKa = 5.65QDD51 pKa = 3.7GAEE54 pKa = 4.43HH55 pKa = 5.95IHH57 pKa = 6.25VFLALTEE64 pKa = 4.54PIRR67 pKa = 11.84IKK69 pKa = 10.84SNEE72 pKa = 3.81MDD74 pKa = 3.25VFDD77 pKa = 5.62IVDD80 pKa = 3.41EE81 pKa = 4.58DD82 pKa = 4.19EE83 pKa = 4.38PDD85 pKa = 3.2KK86 pKa = 11.01RR87 pKa = 11.84VYY89 pKa = 10.09HH90 pKa = 6.55CNIQSVKK97 pKa = 10.29SPKK100 pKa = 10.16DD101 pKa = 3.35AIQYY105 pKa = 7.82VKK107 pKa = 10.94KK108 pKa = 9.7NGHH111 pKa = 6.84FITYY115 pKa = 7.76GICPWKK121 pKa = 10.87VPLTMAEE128 pKa = 4.31KK129 pKa = 11.09NEE131 pKa = 3.88LLQSKK136 pKa = 9.54SLHH139 pKa = 5.75EE140 pKa = 4.09LVEE143 pKa = 4.37SGMVSIFKK151 pKa = 10.32IPQLQKK157 pKa = 10.86AISILQNEE165 pKa = 4.82LGNVEE170 pKa = 4.02RR171 pKa = 11.84QPPKK175 pKa = 8.47VHH177 pKa = 6.78WYY179 pKa = 9.64YY180 pKa = 11.5GSTGTGKK187 pKa = 8.11TRR189 pKa = 11.84TAYY192 pKa = 10.07EE193 pKa = 3.94EE194 pKa = 4.24AKK196 pKa = 10.46EE197 pKa = 4.33SEE199 pKa = 3.88DD200 pKa = 4.42GYY202 pKa = 10.34WISHH206 pKa = 5.23EE207 pKa = 4.1TGEE210 pKa = 4.47WFDD213 pKa = 4.39GYY215 pKa = 9.91IGQGIAILDD224 pKa = 4.7DD225 pKa = 3.05IRR227 pKa = 11.84AATWPLQKK235 pKa = 10.63LLRR238 pKa = 11.84LCDD241 pKa = 3.95RR242 pKa = 11.84YY243 pKa = 10.25PIQVPIKK250 pKa = 10.79GGFTWWRR257 pKa = 11.84PRR259 pKa = 11.84EE260 pKa = 4.03IYY262 pKa = 8.78ITAPGRR268 pKa = 11.84PEE270 pKa = 5.81DD271 pKa = 3.3IYY273 pKa = 11.75KK274 pKa = 10.68NYY276 pKa = 7.75TTGQPYY282 pKa = 11.2DD283 pKa = 3.97NIEE286 pKa = 3.86QLNRR290 pKa = 11.84RR291 pKa = 11.84ITEE294 pKa = 4.14MIDD297 pKa = 3.26FDD299 pKa = 5.54SEE301 pKa = 4.51TEE303 pKa = 4.74PIDD306 pKa = 3.39QTLYY310 pKa = 10.21WKK312 pKa = 10.71GG313 pKa = 3.26
Molecular weight: 36.64 kDa
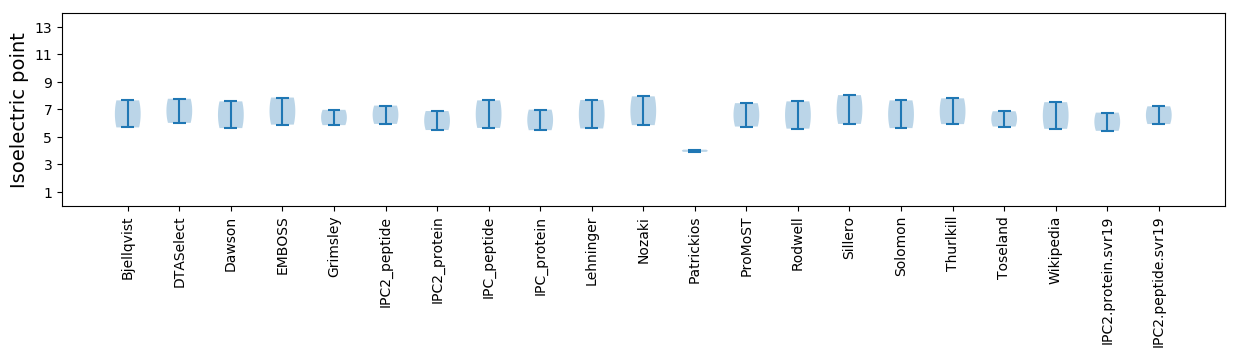
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MFX8|A0A168MFX8_9VIRU Capsid protein OS=Bovine faeces associated circular DNA virus 2 OX=1843765 PE=4 SV=1
MM1 pKa = 8.04PIHH4 pKa = 7.1AIISPKK10 pKa = 8.68FTYY13 pKa = 9.31GQGYY17 pKa = 7.65NHH19 pKa = 5.59VQIRR23 pKa = 11.84VTANSTNAWKK33 pKa = 10.79NGMFDD38 pKa = 3.39YY39 pKa = 10.87SSRR42 pKa = 11.84TDD44 pKa = 2.97TEE46 pKa = 4.27MAQHH50 pKa = 6.55VIMPNIEE57 pKa = 3.99FPVIGNGHH65 pKa = 6.3SNRR68 pKa = 11.84KK69 pKa = 8.65SNRR72 pKa = 11.84LTVSSIRR79 pKa = 11.84VKK81 pKa = 10.69MNFCMDD87 pKa = 4.13GEE89 pKa = 4.54CLRR92 pKa = 11.84KK93 pKa = 9.87FNWFSSAEE101 pKa = 4.24TVTGSSQLPLNPHH114 pKa = 6.76LFLKK118 pKa = 10.61FRR120 pKa = 11.84LFLLSVDD127 pKa = 4.26DD128 pKa = 5.32DD129 pKa = 4.39IEE131 pKa = 4.25VSRR134 pKa = 11.84NKK136 pKa = 10.3LLNWFLATHH145 pKa = 6.1CMYY148 pKa = 10.52RR149 pKa = 11.84YY150 pKa = 6.49PTVEE154 pKa = 4.22EE155 pKa = 4.26VLKK158 pKa = 10.9GPSDD162 pKa = 3.61TAVTDD167 pKa = 3.78TPGPTSVHH175 pKa = 5.92SSVLRR180 pKa = 11.84VTTDD184 pKa = 2.51WTGKK188 pKa = 10.37FNVLADD194 pKa = 3.88RR195 pKa = 11.84KK196 pKa = 9.18FTISAKK202 pKa = 10.35NPQFDD207 pKa = 3.45VDD209 pKa = 3.55MTIPLRR215 pKa = 11.84KK216 pKa = 9.67EE217 pKa = 3.74FVFDD221 pKa = 3.79EE222 pKa = 4.9NYY224 pKa = 10.66DD225 pKa = 3.62ATSKK229 pKa = 10.91LLYY232 pKa = 9.28PKK234 pKa = 10.1LYY236 pKa = 10.88LFILPPLSWEE246 pKa = 3.98VDD248 pKa = 3.54CDD250 pKa = 3.58VMTSYY255 pKa = 10.78FCKK258 pKa = 10.12HH259 pKa = 5.4YY260 pKa = 10.44QSSSAIAPLCFHH272 pKa = 6.84VYY274 pKa = 9.07SWTKK278 pKa = 11.07LNFVDD283 pKa = 5.26LL284 pKa = 4.3
MM1 pKa = 8.04PIHH4 pKa = 7.1AIISPKK10 pKa = 8.68FTYY13 pKa = 9.31GQGYY17 pKa = 7.65NHH19 pKa = 5.59VQIRR23 pKa = 11.84VTANSTNAWKK33 pKa = 10.79NGMFDD38 pKa = 3.39YY39 pKa = 10.87SSRR42 pKa = 11.84TDD44 pKa = 2.97TEE46 pKa = 4.27MAQHH50 pKa = 6.55VIMPNIEE57 pKa = 3.99FPVIGNGHH65 pKa = 6.3SNRR68 pKa = 11.84KK69 pKa = 8.65SNRR72 pKa = 11.84LTVSSIRR79 pKa = 11.84VKK81 pKa = 10.69MNFCMDD87 pKa = 4.13GEE89 pKa = 4.54CLRR92 pKa = 11.84KK93 pKa = 9.87FNWFSSAEE101 pKa = 4.24TVTGSSQLPLNPHH114 pKa = 6.76LFLKK118 pKa = 10.61FRR120 pKa = 11.84LFLLSVDD127 pKa = 4.26DD128 pKa = 5.32DD129 pKa = 4.39IEE131 pKa = 4.25VSRR134 pKa = 11.84NKK136 pKa = 10.3LLNWFLATHH145 pKa = 6.1CMYY148 pKa = 10.52RR149 pKa = 11.84YY150 pKa = 6.49PTVEE154 pKa = 4.22EE155 pKa = 4.26VLKK158 pKa = 10.9GPSDD162 pKa = 3.61TAVTDD167 pKa = 3.78TPGPTSVHH175 pKa = 5.92SSVLRR180 pKa = 11.84VTTDD184 pKa = 2.51WTGKK188 pKa = 10.37FNVLADD194 pKa = 3.88RR195 pKa = 11.84KK196 pKa = 9.18FTISAKK202 pKa = 10.35NPQFDD207 pKa = 3.45VDD209 pKa = 3.55MTIPLRR215 pKa = 11.84KK216 pKa = 9.67EE217 pKa = 3.74FVFDD221 pKa = 3.79EE222 pKa = 4.9NYY224 pKa = 10.66DD225 pKa = 3.62ATSKK229 pKa = 10.91LLYY232 pKa = 9.28PKK234 pKa = 10.1LYY236 pKa = 10.88LFILPPLSWEE246 pKa = 3.98VDD248 pKa = 3.54CDD250 pKa = 3.58VMTSYY255 pKa = 10.78FCKK258 pKa = 10.12HH259 pKa = 5.4YY260 pKa = 10.44QSSSAIAPLCFHH272 pKa = 6.84VYY274 pKa = 9.07SWTKK278 pKa = 11.07LNFVDD283 pKa = 5.26LL284 pKa = 4.3
Molecular weight: 32.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
597 |
284 |
313 |
298.5 |
34.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.02 ± 0.129 | 1.675 ± 0.274 |
5.863 ± 0.077 | 5.863 ± 1.466 |
4.355 ± 1.462 | 4.858 ± 0.837 |
2.848 ± 0.201 | 7.203 ± 1.644 |
6.03 ± 0.248 | 7.873 ± 0.582 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.513 ± 0.411 | 5.193 ± 0.497 |
5.528 ± 0.066 | 4.188 ± 1.299 |
4.69 ± 0.291 | 6.7 ± 1.537 |
6.7 ± 0.655 | 6.365 ± 0.865 |
2.68 ± 0.355 | 4.858 ± 0.616 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
