
BtRs-BetaCoV/HuB2013
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Sarbecovirus; Severe acute respiratory syndrome-related coronavirus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
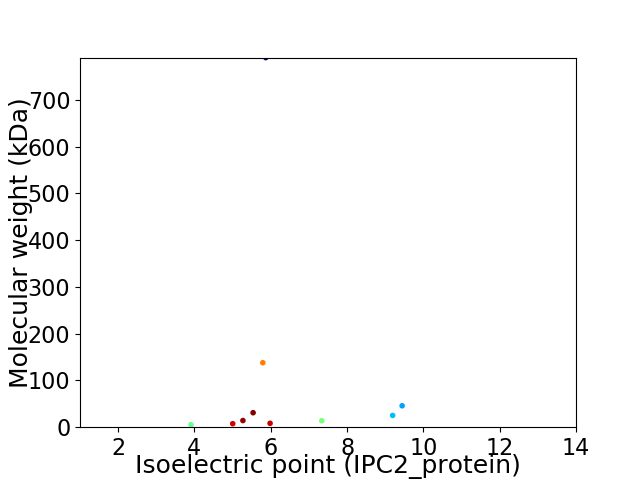
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
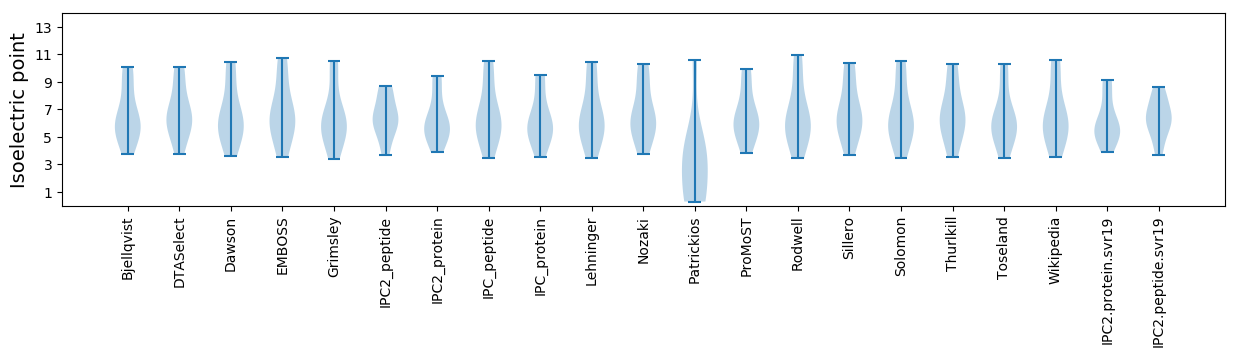
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U1WJY3|A0A0U1WJY3_SARS Accessory protein 7b OS=BtRs-BetaCoV/HuB2013 OX=1503302 PE=4 SV=1
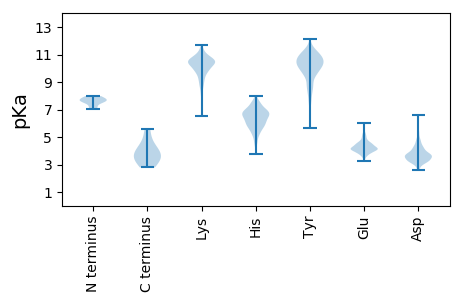
MM1 pKa = 7.71NEE3 pKa = 3.36LTLIDD8 pKa = 5.36FYY10 pKa = 11.69LCFLAFLLFLVLIMLIIFWFSLEE33 pKa = 4.06LQDD36 pKa = 5.95IEE38 pKa = 4.55EE39 pKa = 4.77PCNKK43 pKa = 9.46VV44 pKa = 2.79
MM1 pKa = 7.71NEE3 pKa = 3.36LTLIDD8 pKa = 5.36FYY10 pKa = 11.69LCFLAFLLFLVLIMLIIFWFSLEE33 pKa = 4.06LQDD36 pKa = 5.95IEE38 pKa = 4.55EE39 pKa = 4.77PCNKK43 pKa = 9.46VV44 pKa = 2.79
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U1WHJ2|A0A0U1WHJ2_SARS Membrane protein OS=BtRs-BetaCoV/HuB2013 OX=1503302 GN=M PE=3 SV=1
MM1 pKa = 7.77SDD3 pKa = 4.56NGPQNQRR10 pKa = 11.84SAPRR14 pKa = 11.84ITFGGPSDD22 pKa = 3.8STDD25 pKa = 3.04NNQDD29 pKa = 3.07GGRR32 pKa = 11.84SGARR36 pKa = 11.84PKK38 pKa = 10.25QRR40 pKa = 11.84RR41 pKa = 11.84PQGLPNNTASWFTALTQHH59 pKa = 6.06GKK61 pKa = 10.36EE62 pKa = 3.96EE63 pKa = 4.07LRR65 pKa = 11.84FPRR68 pKa = 11.84GQGVPINTNSGKK80 pKa = 10.01DD81 pKa = 3.58DD82 pKa = 3.56QIGYY86 pKa = 8.17YY87 pKa = 9.92RR88 pKa = 11.84RR89 pKa = 11.84ATRR92 pKa = 11.84RR93 pKa = 11.84VRR95 pKa = 11.84GGDD98 pKa = 3.09GKK100 pKa = 10.39MKK102 pKa = 9.92EE103 pKa = 4.5LSPRR107 pKa = 11.84WYY109 pKa = 10.01FYY111 pKa = 11.5YY112 pKa = 10.73LGTGPEE118 pKa = 3.63ASLPYY123 pKa = 9.9GANKK127 pKa = 9.97EE128 pKa = 4.24GIVWVATEE136 pKa = 4.37GALNTPKK143 pKa = 10.91DD144 pKa = 4.0HH145 pKa = 7.46IGTRR149 pKa = 11.84NPNNNAAIVLQLPQGTTLPKK169 pKa = 10.34GFYY172 pKa = 10.67AEE174 pKa = 4.5GSRR177 pKa = 11.84GGSQASSRR185 pKa = 11.84SSSRR189 pKa = 11.84SRR191 pKa = 11.84GNSRR195 pKa = 11.84NSTPGSSRR203 pKa = 11.84GNSPARR209 pKa = 11.84MASGGGEE216 pKa = 3.84TALALLLLDD225 pKa = 4.89RR226 pKa = 11.84LNQLEE231 pKa = 4.58SKK233 pKa = 10.86VSGKK237 pKa = 10.04GQQQQGQTVTKK248 pKa = 9.98KK249 pKa = 10.44SAAEE253 pKa = 3.73ASKK256 pKa = 10.76KK257 pKa = 9.61PRR259 pKa = 11.84QKK261 pKa = 9.55RR262 pKa = 11.84TATKK266 pKa = 10.22SYY268 pKa = 11.04NVTQAFGRR276 pKa = 11.84RR277 pKa = 11.84GPEE280 pKa = 3.4QTQGNFGDD288 pKa = 3.91QEE290 pKa = 4.91LIRR293 pKa = 11.84QGTDD297 pKa = 2.96YY298 pKa = 11.27KK299 pKa = 10.5HH300 pKa = 6.32WPQIAQFAPSASAFFGMSRR319 pKa = 11.84IGMEE323 pKa = 4.16VTPSGTWLTYY333 pKa = 10.42HH334 pKa = 7.06GAIKK338 pKa = 10.61LDD340 pKa = 4.18DD341 pKa = 4.89KK342 pKa = 11.64DD343 pKa = 3.91PQFKK347 pKa = 11.14DD348 pKa = 3.7NVILLNKK355 pKa = 10.01HH356 pKa = 4.67IDD358 pKa = 3.35AYY360 pKa = 9.77KK361 pKa = 9.88TFPPTEE367 pKa = 3.95PKK369 pKa = 9.88KK370 pKa = 10.6DD371 pKa = 3.48KK372 pKa = 10.73KK373 pKa = 10.99KK374 pKa = 9.88KK375 pKa = 8.29TDD377 pKa = 3.29EE378 pKa = 4.34AQPLPQRR385 pKa = 11.84KK386 pKa = 8.41KK387 pKa = 10.06QPTVTLLPAADD398 pKa = 3.84MDD400 pKa = 4.26DD401 pKa = 5.3FSRR404 pKa = 11.84QLQNSMSGASADD416 pKa = 3.63STQAA420 pKa = 3.04
MM1 pKa = 7.77SDD3 pKa = 4.56NGPQNQRR10 pKa = 11.84SAPRR14 pKa = 11.84ITFGGPSDD22 pKa = 3.8STDD25 pKa = 3.04NNQDD29 pKa = 3.07GGRR32 pKa = 11.84SGARR36 pKa = 11.84PKK38 pKa = 10.25QRR40 pKa = 11.84RR41 pKa = 11.84PQGLPNNTASWFTALTQHH59 pKa = 6.06GKK61 pKa = 10.36EE62 pKa = 3.96EE63 pKa = 4.07LRR65 pKa = 11.84FPRR68 pKa = 11.84GQGVPINTNSGKK80 pKa = 10.01DD81 pKa = 3.58DD82 pKa = 3.56QIGYY86 pKa = 8.17YY87 pKa = 9.92RR88 pKa = 11.84RR89 pKa = 11.84ATRR92 pKa = 11.84RR93 pKa = 11.84VRR95 pKa = 11.84GGDD98 pKa = 3.09GKK100 pKa = 10.39MKK102 pKa = 9.92EE103 pKa = 4.5LSPRR107 pKa = 11.84WYY109 pKa = 10.01FYY111 pKa = 11.5YY112 pKa = 10.73LGTGPEE118 pKa = 3.63ASLPYY123 pKa = 9.9GANKK127 pKa = 9.97EE128 pKa = 4.24GIVWVATEE136 pKa = 4.37GALNTPKK143 pKa = 10.91DD144 pKa = 4.0HH145 pKa = 7.46IGTRR149 pKa = 11.84NPNNNAAIVLQLPQGTTLPKK169 pKa = 10.34GFYY172 pKa = 10.67AEE174 pKa = 4.5GSRR177 pKa = 11.84GGSQASSRR185 pKa = 11.84SSSRR189 pKa = 11.84SRR191 pKa = 11.84GNSRR195 pKa = 11.84NSTPGSSRR203 pKa = 11.84GNSPARR209 pKa = 11.84MASGGGEE216 pKa = 3.84TALALLLLDD225 pKa = 4.89RR226 pKa = 11.84LNQLEE231 pKa = 4.58SKK233 pKa = 10.86VSGKK237 pKa = 10.04GQQQQGQTVTKK248 pKa = 9.98KK249 pKa = 10.44SAAEE253 pKa = 3.73ASKK256 pKa = 10.76KK257 pKa = 9.61PRR259 pKa = 11.84QKK261 pKa = 9.55RR262 pKa = 11.84TATKK266 pKa = 10.22SYY268 pKa = 11.04NVTQAFGRR276 pKa = 11.84RR277 pKa = 11.84GPEE280 pKa = 3.4QTQGNFGDD288 pKa = 3.91QEE290 pKa = 4.91LIRR293 pKa = 11.84QGTDD297 pKa = 2.96YY298 pKa = 11.27KK299 pKa = 10.5HH300 pKa = 6.32WPQIAQFAPSASAFFGMSRR319 pKa = 11.84IGMEE323 pKa = 4.16VTPSGTWLTYY333 pKa = 10.42HH334 pKa = 7.06GAIKK338 pKa = 10.61LDD340 pKa = 4.18DD341 pKa = 4.89KK342 pKa = 11.64DD343 pKa = 3.91PQFKK347 pKa = 11.14DD348 pKa = 3.7NVILLNKK355 pKa = 10.01HH356 pKa = 4.67IDD358 pKa = 3.35AYY360 pKa = 9.77KK361 pKa = 9.88TFPPTEE367 pKa = 3.95PKK369 pKa = 9.88KK370 pKa = 10.6DD371 pKa = 3.48KK372 pKa = 10.73KK373 pKa = 10.99KK374 pKa = 9.88KK375 pKa = 8.29TDD377 pKa = 3.29EE378 pKa = 4.34AQPLPQRR385 pKa = 11.84KK386 pKa = 8.41KK387 pKa = 10.06QPTVTLLPAADD398 pKa = 3.84MDD400 pKa = 4.26DD401 pKa = 5.3FSRR404 pKa = 11.84QLQNSMSGASADD416 pKa = 3.63STQAA420 pKa = 3.04
Molecular weight: 45.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9652 |
44 |
7070 |
965.2 |
107.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.138 ± 0.261 | 3.087 ± 0.488 |
5.232 ± 0.448 | 4.755 ± 0.434 |
4.869 ± 0.407 | 5.968 ± 0.652 |
2.082 ± 0.317 | 5.16 ± 0.867 |
5.595 ± 0.563 | 9.511 ± 0.985 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.352 ± 0.261 | 5.18 ± 0.535 |
4.072 ± 0.434 | 3.751 ± 0.714 |
3.823 ± 0.595 | 6.714 ± 0.567 |
7.035 ± 0.242 | 7.967 ± 0.864 |
1.129 ± 0.196 | 4.579 ± 0.315 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
