
Gopherus associated genomovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus haeme2
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
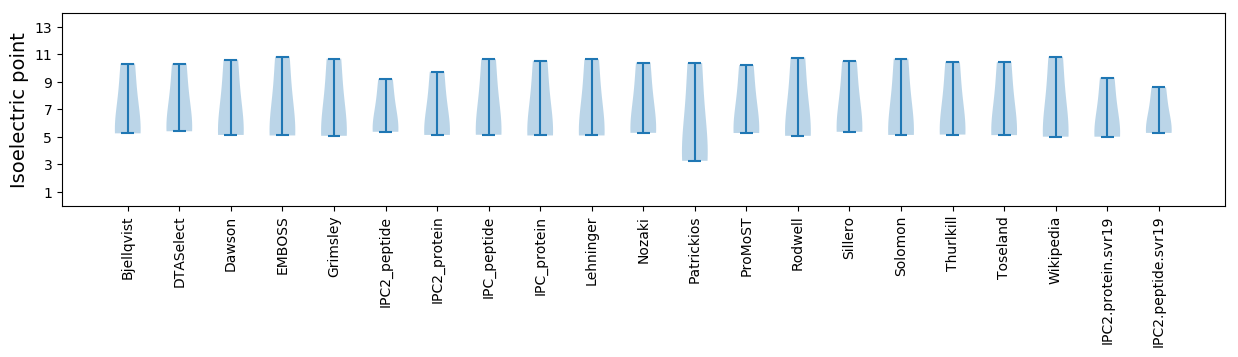
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A384X6V0|A0A384X6V0_9VIRU Replication-associated protein OS=Gopherus associated genomovirus 1 OX=2041417 PE=3 SV=1
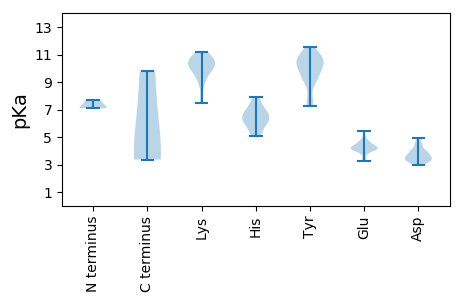
MM1 pKa = 7.1FRR3 pKa = 11.84FAARR7 pKa = 11.84YY8 pKa = 9.22GLLTYY13 pKa = 7.26PQCGDD18 pKa = 3.79LDD20 pKa = 3.44PWAIVEE26 pKa = 4.05MLGRR30 pKa = 11.84LGAEE34 pKa = 4.08CIVGRR39 pKa = 11.84EE40 pKa = 4.01SHH42 pKa = 6.15QDD44 pKa = 3.23GGVHH48 pKa = 5.67LHH50 pKa = 6.33AFFMFEE56 pKa = 4.48SKK58 pKa = 10.78FEE60 pKa = 4.17SRR62 pKa = 11.84NVRR65 pKa = 11.84IFDD68 pKa = 3.59VDD70 pKa = 3.42GMHH73 pKa = 6.97PNVVRR78 pKa = 11.84GYY80 pKa = 7.6STPEE84 pKa = 3.24KK85 pKa = 10.21GYY87 pKa = 11.0AYY89 pKa = 10.04AIKK92 pKa = 10.8DD93 pKa = 3.37GDD95 pKa = 4.03FVAGGLGMEE104 pKa = 5.06SIRR107 pKa = 11.84RR108 pKa = 11.84PVSEE112 pKa = 4.73SRR114 pKa = 11.84SVWDD118 pKa = 4.44EE119 pKa = 3.5IVLAGSRR126 pKa = 11.84EE127 pKa = 4.11EE128 pKa = 4.37FFEE131 pKa = 4.29ACARR135 pKa = 11.84LAPRR139 pKa = 11.84ALCCSFTSLNCYY151 pKa = 10.31ADD153 pKa = 3.12WKK155 pKa = 10.2YY156 pKa = 10.85RR157 pKa = 11.84PEE159 pKa = 4.19PVPYY163 pKa = 9.1EE164 pKa = 3.96HH165 pKa = 7.62PGNVSLSTANFPEE178 pKa = 4.01LAAWVSHH185 pKa = 5.1NLEE188 pKa = 4.37GLEE191 pKa = 4.65HH192 pKa = 7.05PGKK195 pKa = 10.23CPPSAEE201 pKa = 4.07EE202 pKa = 3.67QILAIFLPTLCYY214 pKa = 10.52GG215 pKa = 3.65
MM1 pKa = 7.1FRR3 pKa = 11.84FAARR7 pKa = 11.84YY8 pKa = 9.22GLLTYY13 pKa = 7.26PQCGDD18 pKa = 3.79LDD20 pKa = 3.44PWAIVEE26 pKa = 4.05MLGRR30 pKa = 11.84LGAEE34 pKa = 4.08CIVGRR39 pKa = 11.84EE40 pKa = 4.01SHH42 pKa = 6.15QDD44 pKa = 3.23GGVHH48 pKa = 5.67LHH50 pKa = 6.33AFFMFEE56 pKa = 4.48SKK58 pKa = 10.78FEE60 pKa = 4.17SRR62 pKa = 11.84NVRR65 pKa = 11.84IFDD68 pKa = 3.59VDD70 pKa = 3.42GMHH73 pKa = 6.97PNVVRR78 pKa = 11.84GYY80 pKa = 7.6STPEE84 pKa = 3.24KK85 pKa = 10.21GYY87 pKa = 11.0AYY89 pKa = 10.04AIKK92 pKa = 10.8DD93 pKa = 3.37GDD95 pKa = 4.03FVAGGLGMEE104 pKa = 5.06SIRR107 pKa = 11.84RR108 pKa = 11.84PVSEE112 pKa = 4.73SRR114 pKa = 11.84SVWDD118 pKa = 4.44EE119 pKa = 3.5IVLAGSRR126 pKa = 11.84EE127 pKa = 4.11EE128 pKa = 4.37FFEE131 pKa = 4.29ACARR135 pKa = 11.84LAPRR139 pKa = 11.84ALCCSFTSLNCYY151 pKa = 10.31ADD153 pKa = 3.12WKK155 pKa = 10.2YY156 pKa = 10.85RR157 pKa = 11.84PEE159 pKa = 4.19PVPYY163 pKa = 9.1EE164 pKa = 3.96HH165 pKa = 7.62PGNVSLSTANFPEE178 pKa = 4.01LAAWVSHH185 pKa = 5.1NLEE188 pKa = 4.37GLEE191 pKa = 4.65HH192 pKa = 7.05PGKK195 pKa = 10.23CPPSAEE201 pKa = 4.07EE202 pKa = 3.67QILAIFLPTLCYY214 pKa = 10.52GG215 pKa = 3.65
Molecular weight: 24.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A384X6W5|A0A384X6W5_9VIRU Capsid protein OS=Gopherus associated genomovirus 1 OX=2041417 PE=4 SV=1
MM1 pKa = 7.68AYY3 pKa = 10.13GRR5 pKa = 11.84RR6 pKa = 11.84SYY8 pKa = 11.16RR9 pKa = 11.84RR10 pKa = 11.84TTTRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 9.36RR17 pKa = 11.84AGSSRR22 pKa = 11.84RR23 pKa = 11.84SYY25 pKa = 10.58RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 7.6TYY30 pKa = 7.57TRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84TYY36 pKa = 10.14GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84MSTRR44 pKa = 11.84TVLNKK49 pKa = 9.7TSQKK53 pKa = 10.44KK54 pKa = 9.08RR55 pKa = 11.84DD56 pKa = 3.51NMLAYY61 pKa = 10.94SNTTFDD67 pKa = 3.97NPFATTYY74 pKa = 9.5TLGGAIMRR82 pKa = 11.84FPVGNTSIPSEE93 pKa = 4.72FIYY96 pKa = 10.53VWNATGRR103 pKa = 11.84PGEE106 pKa = 4.2NSDD109 pKa = 3.54GQRR112 pKa = 11.84GSKK115 pKa = 10.41LDD117 pKa = 3.38TSLRR121 pKa = 11.84TSEE124 pKa = 4.25TVFARR129 pKa = 11.84GLKK132 pKa = 10.09EE133 pKa = 4.66KK134 pKa = 9.54ITLEE138 pKa = 4.33TNNAAPWHH146 pKa = 5.36WRR148 pKa = 11.84RR149 pKa = 11.84ICFTSKK155 pKa = 11.17DD156 pKa = 3.99DD157 pKa = 4.66FGEE160 pKa = 4.53ADD162 pKa = 4.63PDD164 pKa = 3.38TSAYY168 pKa = 9.37FRR170 pKa = 11.84RR171 pKa = 11.84TSNGRR176 pKa = 11.84VRR178 pKa = 11.84LLRR181 pKa = 11.84AQSDD185 pKa = 4.02SEE187 pKa = 4.27YY188 pKa = 11.17LNDD191 pKa = 3.69EE192 pKa = 4.22LFEE195 pKa = 4.29GEE197 pKa = 5.43RR198 pKa = 11.84NVDD201 pKa = 2.94WLSALNAPLSRR212 pKa = 11.84KK213 pKa = 9.32HH214 pKa = 6.76FSIKK218 pKa = 9.64YY219 pKa = 9.32DD220 pKa = 3.16KK221 pKa = 10.97TRR223 pKa = 11.84VLRR226 pKa = 11.84SQNQAGNIFTFNLWHH241 pKa = 6.37NMGHH245 pKa = 6.2NIVYY249 pKa = 10.32AGEE252 pKa = 4.14QTGEE256 pKa = 4.09RR257 pKa = 11.84MVDD260 pKa = 3.38SAVSVSGRR268 pKa = 11.84PGMGNYY274 pKa = 9.69YY275 pKa = 10.5VVDD278 pKa = 3.87IFRR281 pKa = 11.84KK282 pKa = 10.09HH283 pKa = 6.63GINDD287 pKa = 3.71DD288 pKa = 3.55QSTLTFTPEE297 pKa = 3.46ATFFWHH303 pKa = 6.68EE304 pKa = 4.05KK305 pKa = 9.79
MM1 pKa = 7.68AYY3 pKa = 10.13GRR5 pKa = 11.84RR6 pKa = 11.84SYY8 pKa = 11.16RR9 pKa = 11.84RR10 pKa = 11.84TTTRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 9.36RR17 pKa = 11.84AGSSRR22 pKa = 11.84RR23 pKa = 11.84SYY25 pKa = 10.58RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 7.6TYY30 pKa = 7.57TRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84TYY36 pKa = 10.14GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84MSTRR44 pKa = 11.84TVLNKK49 pKa = 9.7TSQKK53 pKa = 10.44KK54 pKa = 9.08RR55 pKa = 11.84DD56 pKa = 3.51NMLAYY61 pKa = 10.94SNTTFDD67 pKa = 3.97NPFATTYY74 pKa = 9.5TLGGAIMRR82 pKa = 11.84FPVGNTSIPSEE93 pKa = 4.72FIYY96 pKa = 10.53VWNATGRR103 pKa = 11.84PGEE106 pKa = 4.2NSDD109 pKa = 3.54GQRR112 pKa = 11.84GSKK115 pKa = 10.41LDD117 pKa = 3.38TSLRR121 pKa = 11.84TSEE124 pKa = 4.25TVFARR129 pKa = 11.84GLKK132 pKa = 10.09EE133 pKa = 4.66KK134 pKa = 9.54ITLEE138 pKa = 4.33TNNAAPWHH146 pKa = 5.36WRR148 pKa = 11.84RR149 pKa = 11.84ICFTSKK155 pKa = 11.17DD156 pKa = 3.99DD157 pKa = 4.66FGEE160 pKa = 4.53ADD162 pKa = 4.63PDD164 pKa = 3.38TSAYY168 pKa = 9.37FRR170 pKa = 11.84RR171 pKa = 11.84TSNGRR176 pKa = 11.84VRR178 pKa = 11.84LLRR181 pKa = 11.84AQSDD185 pKa = 4.02SEE187 pKa = 4.27YY188 pKa = 11.17LNDD191 pKa = 3.69EE192 pKa = 4.22LFEE195 pKa = 4.29GEE197 pKa = 5.43RR198 pKa = 11.84NVDD201 pKa = 2.94WLSALNAPLSRR212 pKa = 11.84KK213 pKa = 9.32HH214 pKa = 6.76FSIKK218 pKa = 9.64YY219 pKa = 9.32DD220 pKa = 3.16KK221 pKa = 10.97TRR223 pKa = 11.84VLRR226 pKa = 11.84SQNQAGNIFTFNLWHH241 pKa = 6.37NMGHH245 pKa = 6.2NIVYY249 pKa = 10.32AGEE252 pKa = 4.14QTGEE256 pKa = 4.09RR257 pKa = 11.84MVDD260 pKa = 3.38SAVSVSGRR268 pKa = 11.84PGMGNYY274 pKa = 9.69YY275 pKa = 10.5VVDD278 pKa = 3.87IFRR281 pKa = 11.84KK282 pKa = 10.09HH283 pKa = 6.63GINDD287 pKa = 3.71DD288 pKa = 3.55QSTLTFTPEE297 pKa = 3.46ATFFWHH303 pKa = 6.68EE304 pKa = 4.05KK305 pKa = 9.79
Molecular weight: 35.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
845 |
215 |
325 |
281.7 |
32.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.982 ± 0.643 | 1.893 ± 0.753 |
5.444 ± 0.499 | 6.864 ± 1.01 |
5.917 ± 0.304 | 8.639 ± 0.655 |
2.722 ± 0.328 | 3.432 ± 0.098 |
3.669 ± 0.493 | 7.456 ± 0.815 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.249 ± 0.042 | 4.26 ± 0.993 |
4.734 ± 0.885 | 1.775 ± 0.226 |
8.639 ± 1.65 | 7.811 ± 0.344 |
5.207 ± 2.141 | 5.68 ± 0.613 |
2.13 ± 0.148 | 4.497 ± 0.101 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
