
Halieaceae bacterium IMCC3088
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Aequoribacter
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
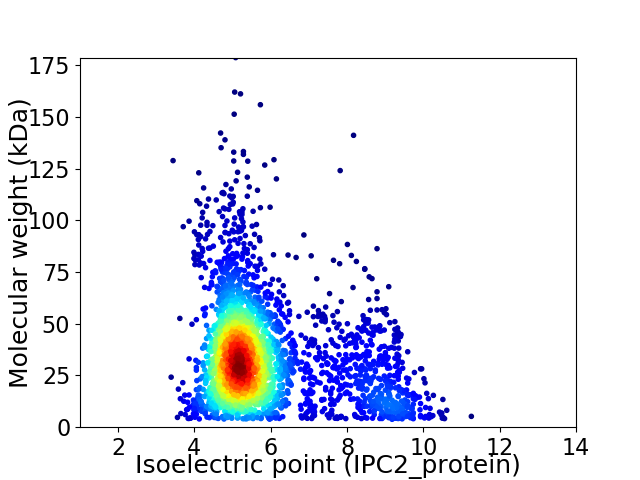
Virtual 2D-PAGE plot for 2855 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F3L401|F3L401_9GAMM GMP synthase [glutamine-hydrolyzing] OS=Halieaceae bacterium IMCC3088 OX=2518989 GN=guaA PE=3 SV=1
MM1 pKa = 7.65RR2 pKa = 11.84AIFLIFVGLLHH13 pKa = 7.28AYY15 pKa = 9.09SANAQEE21 pKa = 4.59AFVLGEE27 pKa = 3.97GRR29 pKa = 11.84LGEE32 pKa = 4.48SVWASDD38 pKa = 5.06LDD40 pKa = 3.98GDD42 pKa = 5.73LIADD46 pKa = 4.5NKK48 pKa = 10.98DD49 pKa = 3.32QFPNAPIGNLTDD61 pKa = 3.46TDD63 pKa = 4.21ADD65 pKa = 3.99GAPNNCDD72 pKa = 3.16QACIDD77 pKa = 3.28SGMRR81 pKa = 11.84ADD83 pKa = 5.82ADD85 pKa = 3.9DD86 pKa = 5.85DD87 pKa = 4.37NDD89 pKa = 3.87GVADD93 pKa = 4.14SEE95 pKa = 5.03DD96 pKa = 4.19AFPLDD101 pKa = 3.35PAEE104 pKa = 4.35TVDD107 pKa = 3.68TDD109 pKa = 4.75GDD111 pKa = 4.2GTGNNADD118 pKa = 4.92LDD120 pKa = 4.41DD121 pKa = 6.16DD122 pKa = 4.98GDD124 pKa = 4.26GYY126 pKa = 11.64ADD128 pKa = 4.16NLDD131 pKa = 4.4AFPIDD136 pKa = 3.9PLEE139 pKa = 4.93HH140 pKa = 7.03SDD142 pKa = 4.91NDD144 pKa = 3.77DD145 pKa = 4.32DD146 pKa = 6.4GIGDD150 pKa = 3.98VADD153 pKa = 5.52DD154 pKa = 5.42DD155 pKa = 5.3DD156 pKa = 6.71DD157 pKa = 5.65NDD159 pKa = 5.4AITDD163 pKa = 3.59ADD165 pKa = 4.51EE166 pKa = 4.38ILLGRR171 pKa = 11.84SPTRR175 pKa = 11.84PEE177 pKa = 4.02YY178 pKa = 11.14VFSTDD183 pKa = 3.1NQSASICLLDD193 pKa = 3.68GEE195 pKa = 5.12RR196 pKa = 11.84IPRR199 pKa = 11.84CWGSNSYY206 pKa = 10.67GQIQPPTSTAFKK218 pKa = 10.28QISLGEE224 pKa = 3.92RR225 pKa = 11.84HH226 pKa = 6.03GCGITLSDD234 pKa = 4.56TVSCWGDD241 pKa = 3.19NAYY244 pKa = 10.21GQASPPEE251 pKa = 4.14NLDD254 pKa = 3.22KK255 pKa = 11.33VKK257 pKa = 10.7QLTSANRR264 pKa = 11.84YY265 pKa = 9.13SCALLEE271 pKa = 5.21DD272 pKa = 5.11DD273 pKa = 6.49SITCWGGKK281 pKa = 10.29YY282 pKa = 10.48YY283 pKa = 10.14MNPLTFSGAKK293 pKa = 9.82KK294 pKa = 9.9IVSGWDD300 pKa = 3.32HH301 pKa = 6.65SCVLHH306 pKa = 6.78SGGISCFGSTGSEE319 pKa = 4.03QIPSDD324 pKa = 3.6LSGSYY329 pKa = 10.44TDD331 pKa = 3.04VHH333 pKa = 6.34AANDD337 pKa = 4.33FVCSEE342 pKa = 4.83DD343 pKa = 3.61STGSTSCYY351 pKa = 10.47GIGKK355 pKa = 9.56DD356 pKa = 2.86IYY358 pKa = 11.08YY359 pKa = 10.63DD360 pKa = 3.54VTSEE364 pKa = 4.19TNINNLTVGSYY375 pKa = 10.36NYY377 pKa = 9.92CSHH380 pKa = 7.01EE381 pKa = 4.07YY382 pKa = 10.82GYY384 pKa = 11.72GLACGGSSSFDD395 pKa = 3.03ILTPPLLNARR405 pKa = 11.84IGTLALTTTMACVLTSTGVTCWGTGANDD433 pKa = 3.38APEE436 pKa = 4.06ILFDD440 pKa = 4.2NDD442 pKa = 2.74QDD444 pKa = 4.01GVNDD448 pKa = 3.96IEE450 pKa = 5.43DD451 pKa = 4.52AFPLDD456 pKa = 3.52AAEE459 pKa = 6.47SIDD462 pKa = 3.83TDD464 pKa = 3.56NDD466 pKa = 4.78GIGNNTDD473 pKa = 4.44LDD475 pKa = 4.43DD476 pKa = 6.4DD477 pKa = 4.27NDD479 pKa = 4.63GYY481 pKa = 11.62SDD483 pKa = 3.87AYY485 pKa = 10.01EE486 pKa = 4.28LEE488 pKa = 4.71KK489 pKa = 10.94GTDD492 pKa = 3.6PLDD495 pKa = 3.81ATDD498 pKa = 6.05FKK500 pKa = 11.17PLYY503 pKa = 9.99DD504 pKa = 4.35DD505 pKa = 4.83LNGIVYY511 pKa = 9.41HH512 pKa = 6.66WSQHH516 pKa = 4.51SLMNAAEE523 pKa = 4.02IHH525 pKa = 6.58RR526 pKa = 11.84ISDD529 pKa = 3.92AEE531 pKa = 4.39VKK533 pKa = 10.54SATNAQGRR541 pKa = 11.84YY542 pKa = 8.69RR543 pKa = 11.84FEE545 pKa = 4.27DD546 pKa = 3.91TPEE549 pKa = 3.9GNYY552 pKa = 9.93EE553 pKa = 4.1LSASQAVTDD562 pKa = 4.0ADD564 pKa = 3.49INRR567 pKa = 11.84TITSADD573 pKa = 3.05ALAALKK579 pKa = 10.3IAVGLNPNSDD589 pKa = 3.8PDD591 pKa = 3.94GDD593 pKa = 4.37GPLEE597 pKa = 4.15ALAVSPYY604 pKa = 10.64QLISADD610 pKa = 3.64MNQDD614 pKa = 2.96GRR616 pKa = 11.84VTSADD621 pKa = 2.99ALAILKK627 pKa = 9.75VAVGLSDD634 pKa = 4.17ALEE637 pKa = 4.42PSWKK641 pKa = 10.15LVADD645 pKa = 4.29AQSLWSTHH653 pKa = 4.97GDD655 pKa = 3.03KK656 pKa = 11.57SNVFDD661 pKa = 4.36ASKK664 pKa = 10.73AYY666 pKa = 10.82LLTYY670 pKa = 9.58PDD672 pKa = 3.34KK673 pKa = 11.38TEE675 pKa = 3.95VNFAAILLGDD685 pKa = 4.48VNPTWKK691 pKa = 9.4PQEE694 pKa = 4.08GTEE697 pKa = 4.4SLNHH701 pKa = 6.42DD702 pKa = 4.56HH703 pKa = 7.31FSMYY707 pKa = 10.76AKK709 pKa = 10.68ASGAPLSLWGIRR721 pKa = 11.84DD722 pKa = 3.41SDD724 pKa = 3.89EE725 pKa = 5.75DD726 pKa = 3.95GLSDD730 pKa = 3.99EE731 pKa = 4.66QEE733 pKa = 4.28EE734 pKa = 4.59TLGTSPLEE742 pKa = 4.41ADD744 pKa = 3.36TDD746 pKa = 4.02ADD748 pKa = 3.83GVNDD752 pKa = 3.74VDD754 pKa = 6.3DD755 pKa = 5.78LFPLDD760 pKa = 4.6PDD762 pKa = 3.8KK763 pKa = 11.62SDD765 pKa = 5.33DD766 pKa = 3.67IPVGAEE772 pKa = 3.86PEE774 pKa = 4.27PVLLASPMKK783 pKa = 10.66ANTSAITMVKK793 pKa = 9.8PVLLRR798 pKa = 11.84GDD800 pKa = 3.61MNDD803 pKa = 3.02WGVSDD808 pKa = 4.67TFDD811 pKa = 3.53EE812 pKa = 5.13LDD814 pKa = 3.98DD815 pKa = 3.91GTYY818 pKa = 9.97QLVMKK823 pKa = 7.21LTPGTYY829 pKa = 7.71TFKK832 pKa = 10.74VASANWSIMDD842 pKa = 4.95LGAQTEE848 pKa = 4.28THH850 pKa = 6.62RR851 pKa = 11.84LIKK854 pKa = 10.58PEE856 pKa = 4.02VPTLLSSEE864 pKa = 4.18PSLPYY869 pKa = 10.59VLVLINEE876 pKa = 4.21TTIVFKK882 pKa = 10.44LTTGEE887 pKa = 3.95HH888 pKa = 4.89KK889 pKa = 10.37KK890 pKa = 11.1YY891 pKa = 10.72YY892 pKa = 8.57ITVFNLPDD900 pKa = 3.65TYY902 pKa = 11.22DD903 pKa = 3.54GPP905 pKa = 4.24
MM1 pKa = 7.65RR2 pKa = 11.84AIFLIFVGLLHH13 pKa = 7.28AYY15 pKa = 9.09SANAQEE21 pKa = 4.59AFVLGEE27 pKa = 3.97GRR29 pKa = 11.84LGEE32 pKa = 4.48SVWASDD38 pKa = 5.06LDD40 pKa = 3.98GDD42 pKa = 5.73LIADD46 pKa = 4.5NKK48 pKa = 10.98DD49 pKa = 3.32QFPNAPIGNLTDD61 pKa = 3.46TDD63 pKa = 4.21ADD65 pKa = 3.99GAPNNCDD72 pKa = 3.16QACIDD77 pKa = 3.28SGMRR81 pKa = 11.84ADD83 pKa = 5.82ADD85 pKa = 3.9DD86 pKa = 5.85DD87 pKa = 4.37NDD89 pKa = 3.87GVADD93 pKa = 4.14SEE95 pKa = 5.03DD96 pKa = 4.19AFPLDD101 pKa = 3.35PAEE104 pKa = 4.35TVDD107 pKa = 3.68TDD109 pKa = 4.75GDD111 pKa = 4.2GTGNNADD118 pKa = 4.92LDD120 pKa = 4.41DD121 pKa = 6.16DD122 pKa = 4.98GDD124 pKa = 4.26GYY126 pKa = 11.64ADD128 pKa = 4.16NLDD131 pKa = 4.4AFPIDD136 pKa = 3.9PLEE139 pKa = 4.93HH140 pKa = 7.03SDD142 pKa = 4.91NDD144 pKa = 3.77DD145 pKa = 4.32DD146 pKa = 6.4GIGDD150 pKa = 3.98VADD153 pKa = 5.52DD154 pKa = 5.42DD155 pKa = 5.3DD156 pKa = 6.71DD157 pKa = 5.65NDD159 pKa = 5.4AITDD163 pKa = 3.59ADD165 pKa = 4.51EE166 pKa = 4.38ILLGRR171 pKa = 11.84SPTRR175 pKa = 11.84PEE177 pKa = 4.02YY178 pKa = 11.14VFSTDD183 pKa = 3.1NQSASICLLDD193 pKa = 3.68GEE195 pKa = 5.12RR196 pKa = 11.84IPRR199 pKa = 11.84CWGSNSYY206 pKa = 10.67GQIQPPTSTAFKK218 pKa = 10.28QISLGEE224 pKa = 3.92RR225 pKa = 11.84HH226 pKa = 6.03GCGITLSDD234 pKa = 4.56TVSCWGDD241 pKa = 3.19NAYY244 pKa = 10.21GQASPPEE251 pKa = 4.14NLDD254 pKa = 3.22KK255 pKa = 11.33VKK257 pKa = 10.7QLTSANRR264 pKa = 11.84YY265 pKa = 9.13SCALLEE271 pKa = 5.21DD272 pKa = 5.11DD273 pKa = 6.49SITCWGGKK281 pKa = 10.29YY282 pKa = 10.48YY283 pKa = 10.14MNPLTFSGAKK293 pKa = 9.82KK294 pKa = 9.9IVSGWDD300 pKa = 3.32HH301 pKa = 6.65SCVLHH306 pKa = 6.78SGGISCFGSTGSEE319 pKa = 4.03QIPSDD324 pKa = 3.6LSGSYY329 pKa = 10.44TDD331 pKa = 3.04VHH333 pKa = 6.34AANDD337 pKa = 4.33FVCSEE342 pKa = 4.83DD343 pKa = 3.61STGSTSCYY351 pKa = 10.47GIGKK355 pKa = 9.56DD356 pKa = 2.86IYY358 pKa = 11.08YY359 pKa = 10.63DD360 pKa = 3.54VTSEE364 pKa = 4.19TNINNLTVGSYY375 pKa = 10.36NYY377 pKa = 9.92CSHH380 pKa = 7.01EE381 pKa = 4.07YY382 pKa = 10.82GYY384 pKa = 11.72GLACGGSSSFDD395 pKa = 3.03ILTPPLLNARR405 pKa = 11.84IGTLALTTTMACVLTSTGVTCWGTGANDD433 pKa = 3.38APEE436 pKa = 4.06ILFDD440 pKa = 4.2NDD442 pKa = 2.74QDD444 pKa = 4.01GVNDD448 pKa = 3.96IEE450 pKa = 5.43DD451 pKa = 4.52AFPLDD456 pKa = 3.52AAEE459 pKa = 6.47SIDD462 pKa = 3.83TDD464 pKa = 3.56NDD466 pKa = 4.78GIGNNTDD473 pKa = 4.44LDD475 pKa = 4.43DD476 pKa = 6.4DD477 pKa = 4.27NDD479 pKa = 4.63GYY481 pKa = 11.62SDD483 pKa = 3.87AYY485 pKa = 10.01EE486 pKa = 4.28LEE488 pKa = 4.71KK489 pKa = 10.94GTDD492 pKa = 3.6PLDD495 pKa = 3.81ATDD498 pKa = 6.05FKK500 pKa = 11.17PLYY503 pKa = 9.99DD504 pKa = 4.35DD505 pKa = 4.83LNGIVYY511 pKa = 9.41HH512 pKa = 6.66WSQHH516 pKa = 4.51SLMNAAEE523 pKa = 4.02IHH525 pKa = 6.58RR526 pKa = 11.84ISDD529 pKa = 3.92AEE531 pKa = 4.39VKK533 pKa = 10.54SATNAQGRR541 pKa = 11.84YY542 pKa = 8.69RR543 pKa = 11.84FEE545 pKa = 4.27DD546 pKa = 3.91TPEE549 pKa = 3.9GNYY552 pKa = 9.93EE553 pKa = 4.1LSASQAVTDD562 pKa = 4.0ADD564 pKa = 3.49INRR567 pKa = 11.84TITSADD573 pKa = 3.05ALAALKK579 pKa = 10.3IAVGLNPNSDD589 pKa = 3.8PDD591 pKa = 3.94GDD593 pKa = 4.37GPLEE597 pKa = 4.15ALAVSPYY604 pKa = 10.64QLISADD610 pKa = 3.64MNQDD614 pKa = 2.96GRR616 pKa = 11.84VTSADD621 pKa = 2.99ALAILKK627 pKa = 9.75VAVGLSDD634 pKa = 4.17ALEE637 pKa = 4.42PSWKK641 pKa = 10.15LVADD645 pKa = 4.29AQSLWSTHH653 pKa = 4.97GDD655 pKa = 3.03KK656 pKa = 11.57SNVFDD661 pKa = 4.36ASKK664 pKa = 10.73AYY666 pKa = 10.82LLTYY670 pKa = 9.58PDD672 pKa = 3.34KK673 pKa = 11.38TEE675 pKa = 3.95VNFAAILLGDD685 pKa = 4.48VNPTWKK691 pKa = 9.4PQEE694 pKa = 4.08GTEE697 pKa = 4.4SLNHH701 pKa = 6.42DD702 pKa = 4.56HH703 pKa = 7.31FSMYY707 pKa = 10.76AKK709 pKa = 10.68ASGAPLSLWGIRR721 pKa = 11.84DD722 pKa = 3.41SDD724 pKa = 3.89EE725 pKa = 5.75DD726 pKa = 3.95GLSDD730 pKa = 3.99EE731 pKa = 4.66QEE733 pKa = 4.28EE734 pKa = 4.59TLGTSPLEE742 pKa = 4.41ADD744 pKa = 3.36TDD746 pKa = 4.02ADD748 pKa = 3.83GVNDD752 pKa = 3.74VDD754 pKa = 6.3DD755 pKa = 5.78LFPLDD760 pKa = 4.6PDD762 pKa = 3.8KK763 pKa = 11.62SDD765 pKa = 5.33DD766 pKa = 3.67IPVGAEE772 pKa = 3.86PEE774 pKa = 4.27PVLLASPMKK783 pKa = 10.66ANTSAITMVKK793 pKa = 9.8PVLLRR798 pKa = 11.84GDD800 pKa = 3.61MNDD803 pKa = 3.02WGVSDD808 pKa = 4.67TFDD811 pKa = 3.53EE812 pKa = 5.13LDD814 pKa = 3.98DD815 pKa = 3.91GTYY818 pKa = 9.97QLVMKK823 pKa = 7.21LTPGTYY829 pKa = 7.71TFKK832 pKa = 10.74VASANWSIMDD842 pKa = 4.95LGAQTEE848 pKa = 4.28THH850 pKa = 6.62RR851 pKa = 11.84LIKK854 pKa = 10.58PEE856 pKa = 4.02VPTLLSSEE864 pKa = 4.18PSLPYY869 pKa = 10.59VLVLINEE876 pKa = 4.21TTIVFKK882 pKa = 10.44LTTGEE887 pKa = 3.95HH888 pKa = 4.89KK889 pKa = 10.37KK890 pKa = 11.1YY891 pKa = 10.72YY892 pKa = 8.57ITVFNLPDD900 pKa = 3.65TYY902 pKa = 11.22DD903 pKa = 3.54GPP905 pKa = 4.24
Molecular weight: 96.94 kDa
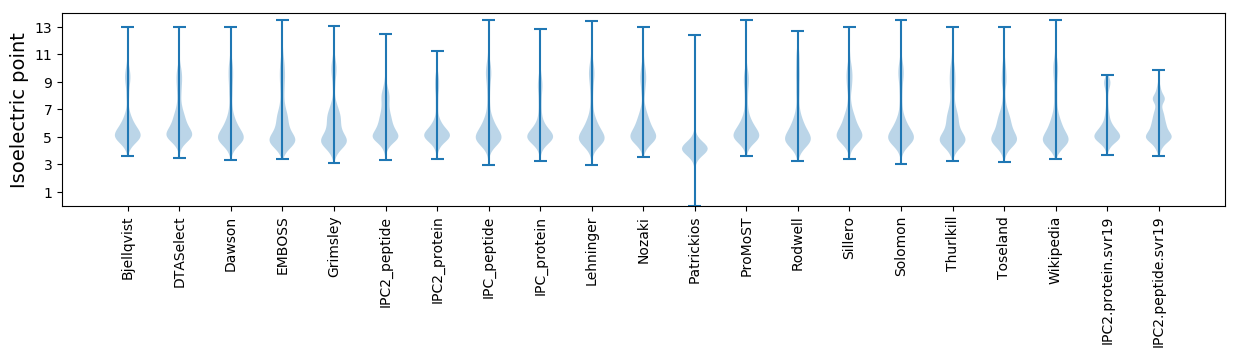
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F3L0U9|F3L0U9_9GAMM Uncharacterized protein OS=Halieaceae bacterium IMCC3088 OX=2518989 GN=IMCC3088_1041 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34SGRR28 pKa = 11.84QLINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34SGRR28 pKa = 11.84QLINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
911031 |
35 |
1596 |
319.1 |
35.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.274 ± 0.044 | 1.011 ± 0.016 |
5.905 ± 0.038 | 6.109 ± 0.039 |
3.829 ± 0.03 | 7.508 ± 0.038 |
2.197 ± 0.025 | 5.577 ± 0.036 |
3.84 ± 0.031 | 10.427 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.449 ± 0.019 | 3.462 ± 0.026 |
4.354 ± 0.024 | 4.268 ± 0.029 |
5.598 ± 0.038 | 6.314 ± 0.038 |
5.381 ± 0.03 | 7.244 ± 0.04 |
1.37 ± 0.019 | 2.882 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
