
Thalassobaculum litoreum DSM 18839
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Thalassobaculaceae; Thalassobaculum; Thalassobaculum litoreum
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
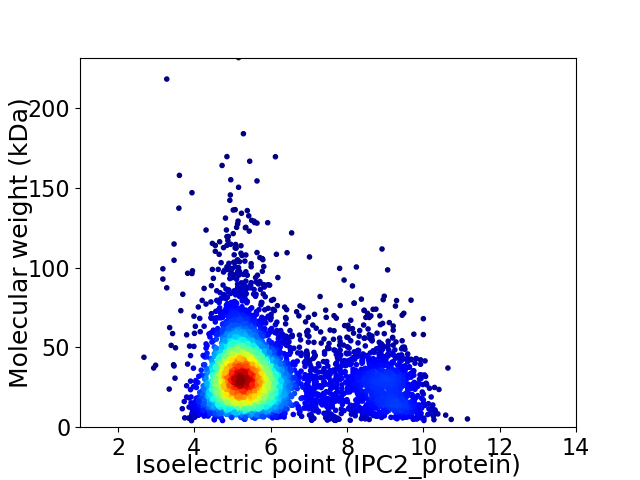
Virtual 2D-PAGE plot for 4959 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G7RJU1|A0A1G7RJU1_9PROT Probable phosphoglycerate mutase OS=Thalassobaculum litoreum DSM 18839 OX=1123362 GN=SAMN05660686_03409 PE=4 SV=1
MM1 pKa = 7.17TVIPSDD7 pKa = 4.29PLFGDD12 pKa = 2.72QWYY15 pKa = 9.28LHH17 pKa = 5.81NTGQSGGTVGQDD29 pKa = 2.68IRR31 pKa = 11.84VLGAWPDD38 pKa = 3.61YY39 pKa = 8.86TGEE42 pKa = 4.44GIRR45 pKa = 11.84IAVIDD50 pKa = 4.09DD51 pKa = 3.66GVRR54 pKa = 11.84YY55 pKa = 6.8THH57 pKa = 7.46PDD59 pKa = 2.84LSGTYY64 pKa = 9.86DD65 pKa = 3.55AEE67 pKa = 4.16NDD69 pKa = 3.66HH70 pKa = 6.98NIVTGEE76 pKa = 3.89GDD78 pKa = 3.45ATPIADD84 pKa = 4.27AYY86 pKa = 10.83SHH88 pKa = 6.21GTQVSGFSAAAANGTGLVGVAYY110 pKa = 9.83GATFTGIRR118 pKa = 11.84ISDD121 pKa = 3.65ADD123 pKa = 3.95GEE125 pKa = 4.66LVNAAPAFHH134 pKa = 6.95LASDD138 pKa = 4.02ADD140 pKa = 3.95VTNNSYY146 pKa = 11.35GNEE149 pKa = 3.73IFYY152 pKa = 10.54EE153 pKa = 4.41EE154 pKa = 5.16PGSLDD159 pKa = 4.27AIADD163 pKa = 3.81TATTGRR169 pKa = 11.84AGLGTVHH176 pKa = 6.28VFASGNSRR184 pKa = 11.84IQGTLSTHH192 pKa = 6.04GPEE195 pKa = 4.27QNSPYY200 pKa = 10.03QITVAATDD208 pKa = 3.56ADD210 pKa = 4.02GQIATFSSPGANVLVAAPGAGVLGVSGEE238 pKa = 4.04DD239 pKa = 3.74DD240 pKa = 5.15GYY242 pKa = 11.48DD243 pKa = 3.56LTDD246 pKa = 3.57GTSFSAPIVSGVAALVLQANPTLGMRR272 pKa = 11.84DD273 pKa = 3.41VQEE276 pKa = 3.83ILAYY280 pKa = 9.9SAYY283 pKa = 9.48DD284 pKa = 3.4TGADD288 pKa = 3.33PLTRR292 pKa = 11.84AEE294 pKa = 4.25ILADD298 pKa = 3.77TVGSEE303 pKa = 3.92AALAEE308 pKa = 4.07LDD310 pKa = 3.9EE311 pKa = 4.73PVRR314 pKa = 11.84AAYY317 pKa = 8.53EE318 pKa = 4.02AGISLALSYY327 pKa = 10.5PWTTVEE333 pKa = 5.66NGATDD338 pKa = 3.44WNGGGLTASHH348 pKa = 7.86DD349 pKa = 3.87YY350 pKa = 11.59GFGQVDD356 pKa = 3.16ARR358 pKa = 11.84AATRR362 pKa = 11.84LAEE365 pKa = 4.29TWDD368 pKa = 3.48ATPNTVTNQAVVTVTSTTAVAVPDD392 pKa = 3.67GDD394 pKa = 3.81AAGVSQSLTVDD405 pKa = 2.67AAMDD409 pKa = 3.73VEE411 pKa = 4.61FATVDD416 pKa = 3.11VDD418 pKa = 4.43IPHH421 pKa = 7.39GDD423 pKa = 3.35LGQLRR428 pKa = 11.84IEE430 pKa = 4.31LTSPHH435 pKa = 6.2GTTSYY440 pKa = 11.13LIYY443 pKa = 11.04NPDD446 pKa = 3.26FEE448 pKa = 6.43AYY450 pKa = 9.44EE451 pKa = 3.73ILARR455 pKa = 11.84QDD457 pKa = 3.25GLLDD461 pKa = 3.98EE462 pKa = 5.93LPDD465 pKa = 3.62LTFFGANVTTLMSSQHH481 pKa = 5.86YY482 pKa = 9.81GEE484 pKa = 4.76SAAGDD489 pKa = 3.21WTLRR493 pKa = 11.84VTDD496 pKa = 4.27TEE498 pKa = 4.42TGTVGTLEE506 pKa = 4.21SWSLTLYY513 pKa = 11.17GDD515 pKa = 3.42ATTADD520 pKa = 3.57DD521 pKa = 4.28RR522 pKa = 11.84YY523 pKa = 11.2VYY525 pKa = 9.83TDD527 pKa = 3.01QYY529 pKa = 10.65ATLATADD536 pKa = 3.54AGRR539 pKa = 11.84SVLVDD544 pKa = 3.36SDD546 pKa = 4.01GGTDD550 pKa = 3.99TINAAAVSGPVAIHH564 pKa = 7.0LDD566 pKa = 3.7GTVGAIYY573 pKa = 10.1GAAFRR578 pKa = 11.84VADD581 pKa = 3.65GTEE584 pKa = 3.65IEE586 pKa = 4.3NLYY589 pKa = 10.07TGDD592 pKa = 4.82GDD594 pKa = 4.97DD595 pKa = 4.85LLAGSALANVLVAGRR610 pKa = 11.84GSDD613 pKa = 3.45SLTGGAGDD621 pKa = 5.55DD622 pKa = 3.95ILDD625 pKa = 4.28GGSGLDD631 pKa = 3.26TARR634 pKa = 11.84FSGLSSSYY642 pKa = 11.02AIGVSDD648 pKa = 4.58SEE650 pKa = 4.79TVVDD654 pKa = 4.45GADD657 pKa = 3.38GRR659 pKa = 11.84DD660 pKa = 3.01ILRR663 pKa = 11.84NTEE666 pKa = 3.58ILAFDD671 pKa = 4.29DD672 pKa = 4.4GNRR675 pKa = 11.84LSTAPVSVSEE685 pKa = 4.5IGFDD689 pKa = 3.58ADD691 pKa = 5.53FYY693 pKa = 11.53LATNDD698 pKa = 3.76DD699 pKa = 3.66VVAAGFDD706 pKa = 3.34SGNAYY711 pKa = 10.43LHH713 pKa = 5.77YY714 pKa = 10.23LQFGGFEE721 pKa = 3.98DD722 pKa = 4.09RR723 pKa = 11.84APNALFDD730 pKa = 4.14SGDD733 pKa = 3.82YY734 pKa = 10.82LAVNADD740 pKa = 3.27VAAAGMNPLLHH751 pKa = 5.53YY752 pKa = 10.16HH753 pKa = 7.19LYY755 pKa = 10.39GHH757 pKa = 7.37LEE759 pKa = 3.86GRR761 pKa = 11.84DD762 pKa = 3.09AGAQFDD768 pKa = 4.34GEE770 pKa = 4.78AYY772 pKa = 10.47LAANPDD778 pKa = 3.38VAGAGIDD785 pKa = 3.73PMLHH789 pKa = 5.8YY790 pKa = 9.78LTVGFGEE797 pKa = 4.4GRR799 pKa = 11.84LFQSDD804 pKa = 4.05PLFGG808 pKa = 4.84
MM1 pKa = 7.17TVIPSDD7 pKa = 4.29PLFGDD12 pKa = 2.72QWYY15 pKa = 9.28LHH17 pKa = 5.81NTGQSGGTVGQDD29 pKa = 2.68IRR31 pKa = 11.84VLGAWPDD38 pKa = 3.61YY39 pKa = 8.86TGEE42 pKa = 4.44GIRR45 pKa = 11.84IAVIDD50 pKa = 4.09DD51 pKa = 3.66GVRR54 pKa = 11.84YY55 pKa = 6.8THH57 pKa = 7.46PDD59 pKa = 2.84LSGTYY64 pKa = 9.86DD65 pKa = 3.55AEE67 pKa = 4.16NDD69 pKa = 3.66HH70 pKa = 6.98NIVTGEE76 pKa = 3.89GDD78 pKa = 3.45ATPIADD84 pKa = 4.27AYY86 pKa = 10.83SHH88 pKa = 6.21GTQVSGFSAAAANGTGLVGVAYY110 pKa = 9.83GATFTGIRR118 pKa = 11.84ISDD121 pKa = 3.65ADD123 pKa = 3.95GEE125 pKa = 4.66LVNAAPAFHH134 pKa = 6.95LASDD138 pKa = 4.02ADD140 pKa = 3.95VTNNSYY146 pKa = 11.35GNEE149 pKa = 3.73IFYY152 pKa = 10.54EE153 pKa = 4.41EE154 pKa = 5.16PGSLDD159 pKa = 4.27AIADD163 pKa = 3.81TATTGRR169 pKa = 11.84AGLGTVHH176 pKa = 6.28VFASGNSRR184 pKa = 11.84IQGTLSTHH192 pKa = 6.04GPEE195 pKa = 4.27QNSPYY200 pKa = 10.03QITVAATDD208 pKa = 3.56ADD210 pKa = 4.02GQIATFSSPGANVLVAAPGAGVLGVSGEE238 pKa = 4.04DD239 pKa = 3.74DD240 pKa = 5.15GYY242 pKa = 11.48DD243 pKa = 3.56LTDD246 pKa = 3.57GTSFSAPIVSGVAALVLQANPTLGMRR272 pKa = 11.84DD273 pKa = 3.41VQEE276 pKa = 3.83ILAYY280 pKa = 9.9SAYY283 pKa = 9.48DD284 pKa = 3.4TGADD288 pKa = 3.33PLTRR292 pKa = 11.84AEE294 pKa = 4.25ILADD298 pKa = 3.77TVGSEE303 pKa = 3.92AALAEE308 pKa = 4.07LDD310 pKa = 3.9EE311 pKa = 4.73PVRR314 pKa = 11.84AAYY317 pKa = 8.53EE318 pKa = 4.02AGISLALSYY327 pKa = 10.5PWTTVEE333 pKa = 5.66NGATDD338 pKa = 3.44WNGGGLTASHH348 pKa = 7.86DD349 pKa = 3.87YY350 pKa = 11.59GFGQVDD356 pKa = 3.16ARR358 pKa = 11.84AATRR362 pKa = 11.84LAEE365 pKa = 4.29TWDD368 pKa = 3.48ATPNTVTNQAVVTVTSTTAVAVPDD392 pKa = 3.67GDD394 pKa = 3.81AAGVSQSLTVDD405 pKa = 2.67AAMDD409 pKa = 3.73VEE411 pKa = 4.61FATVDD416 pKa = 3.11VDD418 pKa = 4.43IPHH421 pKa = 7.39GDD423 pKa = 3.35LGQLRR428 pKa = 11.84IEE430 pKa = 4.31LTSPHH435 pKa = 6.2GTTSYY440 pKa = 11.13LIYY443 pKa = 11.04NPDD446 pKa = 3.26FEE448 pKa = 6.43AYY450 pKa = 9.44EE451 pKa = 3.73ILARR455 pKa = 11.84QDD457 pKa = 3.25GLLDD461 pKa = 3.98EE462 pKa = 5.93LPDD465 pKa = 3.62LTFFGANVTTLMSSQHH481 pKa = 5.86YY482 pKa = 9.81GEE484 pKa = 4.76SAAGDD489 pKa = 3.21WTLRR493 pKa = 11.84VTDD496 pKa = 4.27TEE498 pKa = 4.42TGTVGTLEE506 pKa = 4.21SWSLTLYY513 pKa = 11.17GDD515 pKa = 3.42ATTADD520 pKa = 3.57DD521 pKa = 4.28RR522 pKa = 11.84YY523 pKa = 11.2VYY525 pKa = 9.83TDD527 pKa = 3.01QYY529 pKa = 10.65ATLATADD536 pKa = 3.54AGRR539 pKa = 11.84SVLVDD544 pKa = 3.36SDD546 pKa = 4.01GGTDD550 pKa = 3.99TINAAAVSGPVAIHH564 pKa = 7.0LDD566 pKa = 3.7GTVGAIYY573 pKa = 10.1GAAFRR578 pKa = 11.84VADD581 pKa = 3.65GTEE584 pKa = 3.65IEE586 pKa = 4.3NLYY589 pKa = 10.07TGDD592 pKa = 4.82GDD594 pKa = 4.97DD595 pKa = 4.85LLAGSALANVLVAGRR610 pKa = 11.84GSDD613 pKa = 3.45SLTGGAGDD621 pKa = 5.55DD622 pKa = 3.95ILDD625 pKa = 4.28GGSGLDD631 pKa = 3.26TARR634 pKa = 11.84FSGLSSSYY642 pKa = 11.02AIGVSDD648 pKa = 4.58SEE650 pKa = 4.79TVVDD654 pKa = 4.45GADD657 pKa = 3.38GRR659 pKa = 11.84DD660 pKa = 3.01ILRR663 pKa = 11.84NTEE666 pKa = 3.58ILAFDD671 pKa = 4.29DD672 pKa = 4.4GNRR675 pKa = 11.84LSTAPVSVSEE685 pKa = 4.5IGFDD689 pKa = 3.58ADD691 pKa = 5.53FYY693 pKa = 11.53LATNDD698 pKa = 3.76DD699 pKa = 3.66VVAAGFDD706 pKa = 3.34SGNAYY711 pKa = 10.43LHH713 pKa = 5.77YY714 pKa = 10.23LQFGGFEE721 pKa = 3.98DD722 pKa = 4.09RR723 pKa = 11.84APNALFDD730 pKa = 4.14SGDD733 pKa = 3.82YY734 pKa = 10.82LAVNADD740 pKa = 3.27VAAAGMNPLLHH751 pKa = 5.53YY752 pKa = 10.16HH753 pKa = 7.19LYY755 pKa = 10.39GHH757 pKa = 7.37LEE759 pKa = 3.86GRR761 pKa = 11.84DD762 pKa = 3.09AGAQFDD768 pKa = 4.34GEE770 pKa = 4.78AYY772 pKa = 10.47LAANPDD778 pKa = 3.38VAGAGIDD785 pKa = 3.73PMLHH789 pKa = 5.8YY790 pKa = 9.78LTVGFGEE797 pKa = 4.4GRR799 pKa = 11.84LFQSDD804 pKa = 4.05PLFGG808 pKa = 4.84
Molecular weight: 83.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7KP24|A0A1G7KP24_9PROT Acetyl-CoA acetyltransferase OS=Thalassobaculum litoreum DSM 18839 OX=1123362 GN=SAMN05660686_01124 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.1QPSVLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.1QPSVLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1586320 |
39 |
2184 |
319.9 |
34.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.571 ± 0.054 | 0.851 ± 0.01 |
6.302 ± 0.035 | 5.894 ± 0.032 |
3.485 ± 0.023 | 8.907 ± 0.038 |
2.017 ± 0.02 | 4.939 ± 0.026 |
2.834 ± 0.031 | 9.932 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.529 ± 0.017 | 2.357 ± 0.02 |
5.233 ± 0.027 | 2.822 ± 0.02 |
7.279 ± 0.036 | 5.189 ± 0.025 |
5.56 ± 0.027 | 7.758 ± 0.028 |
1.364 ± 0.014 | 2.176 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
