
Cherry mottle leaf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Trichovirus
Average proteome isoelectric point is 7.46
Get precalculated fractions of proteins
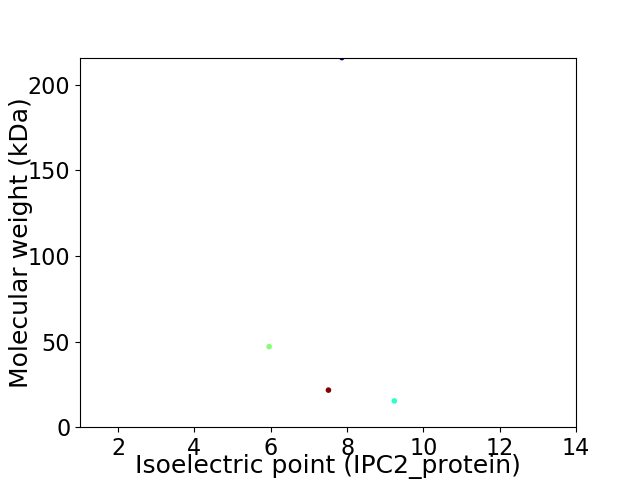
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
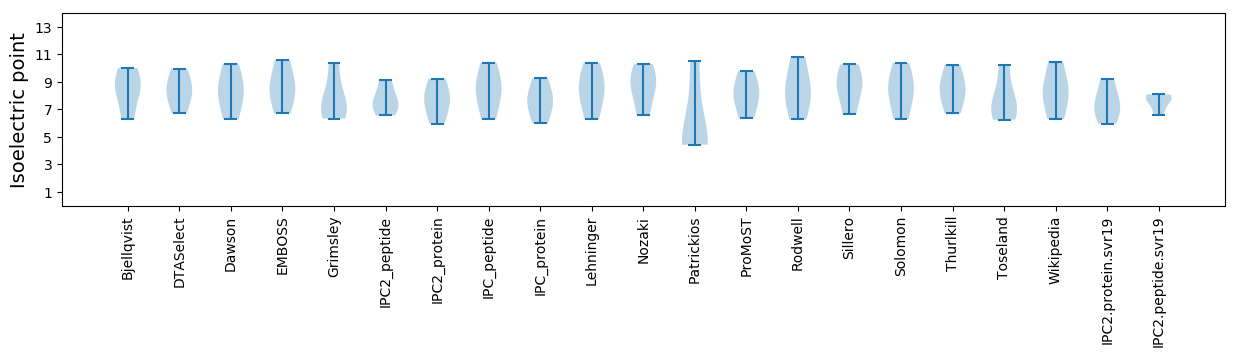
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9IMS9|Q9IMS9_9VIRU 216kDa protein OS=Cherry mottle leaf virus OX=131226 GN=orf1 PE=4 SV=1
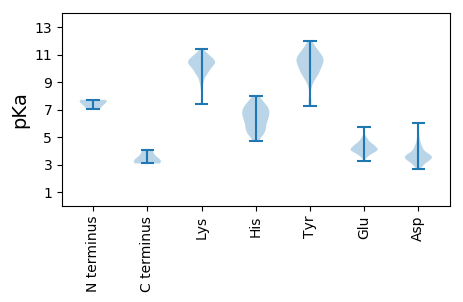
MM1 pKa = 7.05TMMMRR6 pKa = 11.84AHH8 pKa = 7.05KK9 pKa = 10.56SKK11 pKa = 10.24IAEE14 pKa = 3.9GDD16 pKa = 3.52IPISGVKK23 pKa = 9.69SSKK26 pKa = 10.3VYY28 pKa = 11.03SDD30 pKa = 3.11VTPFRR35 pKa = 11.84RR36 pKa = 11.84ASDD39 pKa = 5.08LMIHH43 pKa = 5.83WNEE46 pKa = 3.74FAFKK50 pKa = 10.47IMPDD54 pKa = 4.03DD55 pKa = 4.03IGDD58 pKa = 3.66GGFRR62 pKa = 11.84LMSVPVMPKK71 pKa = 10.51DD72 pKa = 3.63EE73 pKa = 4.46IEE75 pKa = 3.87HH76 pKa = 5.63FRR78 pKa = 11.84RR79 pKa = 11.84EE80 pKa = 4.06RR81 pKa = 11.84GSANYY86 pKa = 7.69VHH88 pKa = 7.23WGALSISIDD97 pKa = 3.52ALFKK101 pKa = 10.45KK102 pKa = 10.54QSGVTGRR109 pKa = 11.84CVVFDD114 pKa = 4.97KK115 pKa = 10.75RR116 pKa = 11.84WEE118 pKa = 4.02NCKK121 pKa = 10.32QSILQTFEE129 pKa = 4.59FNLDD133 pKa = 3.41SGSATMITSPNFSVSLDD150 pKa = 3.71DD151 pKa = 5.3PNLNDD156 pKa = 3.72SLCVVVVFNDD166 pKa = 3.2LNFRR170 pKa = 11.84SEE172 pKa = 4.41SYY174 pKa = 9.79PVSVRR179 pKa = 11.84VGNMCRR185 pKa = 11.84FFDD188 pKa = 3.83SFLGSEE194 pKa = 3.97IFKK197 pKa = 10.91NEE199 pKa = 3.87SNVYY203 pKa = 9.36IDD205 pKa = 4.82SSGADD210 pKa = 3.36LLSYY214 pKa = 10.81KK215 pKa = 10.71SFGFGDD221 pKa = 3.66EE222 pKa = 4.48EE223 pKa = 4.02EE224 pKa = 4.43VEE226 pKa = 4.11RR227 pKa = 11.84LFRR230 pKa = 11.84FAEE233 pKa = 4.29VVPTEE238 pKa = 5.0AIDD241 pKa = 3.55LTVRR245 pKa = 11.84EE246 pKa = 4.4IPGSISNLFRR256 pKa = 11.84KK257 pKa = 9.13KK258 pKa = 10.28RR259 pKa = 11.84VMQYY263 pKa = 11.03GYY265 pKa = 10.81GSKK268 pKa = 10.28SDD270 pKa = 3.43PRR272 pKa = 11.84RR273 pKa = 11.84SKK275 pKa = 10.47GKK277 pKa = 9.61RR278 pKa = 11.84LAGVKK283 pKa = 10.03SLALEE288 pKa = 4.79GEE290 pKa = 4.41LNTYY294 pKa = 9.72LNKK297 pKa = 10.6AGEE300 pKa = 4.43SSNTRR305 pKa = 11.84GVEE308 pKa = 3.91ATHH311 pKa = 6.44SKK313 pKa = 7.4VHH315 pKa = 6.47RR316 pKa = 11.84GKK318 pKa = 10.97GNPFGKK324 pKa = 10.35SKK326 pKa = 10.87DD327 pKa = 3.85SQDD330 pKa = 3.23VGAIEE335 pKa = 5.4SNEE338 pKa = 3.6QDD340 pKa = 3.61TYY342 pKa = 11.47RR343 pKa = 11.84VDD345 pKa = 3.67CLSISSEE352 pKa = 3.81SSTPSKK358 pKa = 10.27IRR360 pKa = 11.84AGKK363 pKa = 9.87EE364 pKa = 3.25VDD366 pKa = 3.4SEE368 pKa = 4.9VNFRR372 pKa = 11.84QHH374 pKa = 6.6CNPRR378 pKa = 11.84DD379 pKa = 3.46VGLNGVYY386 pKa = 10.42FNHH389 pKa = 7.08CSGDD393 pKa = 3.48ASDD396 pKa = 4.41GCGSVSVLQSEE407 pKa = 4.43DD408 pKa = 3.75DD409 pKa = 3.72CWVDD413 pKa = 2.91KK414 pKa = 9.68TFRR417 pKa = 11.84VNKK420 pKa = 10.22RR421 pKa = 11.84GG422 pKa = 3.31
MM1 pKa = 7.05TMMMRR6 pKa = 11.84AHH8 pKa = 7.05KK9 pKa = 10.56SKK11 pKa = 10.24IAEE14 pKa = 3.9GDD16 pKa = 3.52IPISGVKK23 pKa = 9.69SSKK26 pKa = 10.3VYY28 pKa = 11.03SDD30 pKa = 3.11VTPFRR35 pKa = 11.84RR36 pKa = 11.84ASDD39 pKa = 5.08LMIHH43 pKa = 5.83WNEE46 pKa = 3.74FAFKK50 pKa = 10.47IMPDD54 pKa = 4.03DD55 pKa = 4.03IGDD58 pKa = 3.66GGFRR62 pKa = 11.84LMSVPVMPKK71 pKa = 10.51DD72 pKa = 3.63EE73 pKa = 4.46IEE75 pKa = 3.87HH76 pKa = 5.63FRR78 pKa = 11.84RR79 pKa = 11.84EE80 pKa = 4.06RR81 pKa = 11.84GSANYY86 pKa = 7.69VHH88 pKa = 7.23WGALSISIDD97 pKa = 3.52ALFKK101 pKa = 10.45KK102 pKa = 10.54QSGVTGRR109 pKa = 11.84CVVFDD114 pKa = 4.97KK115 pKa = 10.75RR116 pKa = 11.84WEE118 pKa = 4.02NCKK121 pKa = 10.32QSILQTFEE129 pKa = 4.59FNLDD133 pKa = 3.41SGSATMITSPNFSVSLDD150 pKa = 3.71DD151 pKa = 5.3PNLNDD156 pKa = 3.72SLCVVVVFNDD166 pKa = 3.2LNFRR170 pKa = 11.84SEE172 pKa = 4.41SYY174 pKa = 9.79PVSVRR179 pKa = 11.84VGNMCRR185 pKa = 11.84FFDD188 pKa = 3.83SFLGSEE194 pKa = 3.97IFKK197 pKa = 10.91NEE199 pKa = 3.87SNVYY203 pKa = 9.36IDD205 pKa = 4.82SSGADD210 pKa = 3.36LLSYY214 pKa = 10.81KK215 pKa = 10.71SFGFGDD221 pKa = 3.66EE222 pKa = 4.48EE223 pKa = 4.02EE224 pKa = 4.43VEE226 pKa = 4.11RR227 pKa = 11.84LFRR230 pKa = 11.84FAEE233 pKa = 4.29VVPTEE238 pKa = 5.0AIDD241 pKa = 3.55LTVRR245 pKa = 11.84EE246 pKa = 4.4IPGSISNLFRR256 pKa = 11.84KK257 pKa = 9.13KK258 pKa = 10.28RR259 pKa = 11.84VMQYY263 pKa = 11.03GYY265 pKa = 10.81GSKK268 pKa = 10.28SDD270 pKa = 3.43PRR272 pKa = 11.84RR273 pKa = 11.84SKK275 pKa = 10.47GKK277 pKa = 9.61RR278 pKa = 11.84LAGVKK283 pKa = 10.03SLALEE288 pKa = 4.79GEE290 pKa = 4.41LNTYY294 pKa = 9.72LNKK297 pKa = 10.6AGEE300 pKa = 4.43SSNTRR305 pKa = 11.84GVEE308 pKa = 3.91ATHH311 pKa = 6.44SKK313 pKa = 7.4VHH315 pKa = 6.47RR316 pKa = 11.84GKK318 pKa = 10.97GNPFGKK324 pKa = 10.35SKK326 pKa = 10.87DD327 pKa = 3.85SQDD330 pKa = 3.23VGAIEE335 pKa = 5.4SNEE338 pKa = 3.6QDD340 pKa = 3.61TYY342 pKa = 11.47RR343 pKa = 11.84VDD345 pKa = 3.67CLSISSEE352 pKa = 3.81SSTPSKK358 pKa = 10.27IRR360 pKa = 11.84AGKK363 pKa = 9.87EE364 pKa = 3.25VDD366 pKa = 3.4SEE368 pKa = 4.9VNFRR372 pKa = 11.84QHH374 pKa = 6.6CNPRR378 pKa = 11.84DD379 pKa = 3.46VGLNGVYY386 pKa = 10.42FNHH389 pKa = 7.08CSGDD393 pKa = 3.48ASDD396 pKa = 4.41GCGSVSVLQSEE407 pKa = 4.43DD408 pKa = 3.75DD409 pKa = 3.72CWVDD413 pKa = 2.91KK414 pKa = 9.68TFRR417 pKa = 11.84VNKK420 pKa = 10.22RR421 pKa = 11.84GG422 pKa = 3.31
Molecular weight: 47.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9IMS7|Q9IMS7_9VIRU Coat protein OS=Cherry mottle leaf virus OX=131226 GN=orf3 PE=4 SV=1
MM1 pKa = 7.71SFNPKK6 pKa = 9.57NNKK9 pKa = 8.48DD10 pKa = 3.35DD11 pKa = 3.78RR12 pKa = 11.84RR13 pKa = 11.84IFSLCLSMMRR23 pKa = 11.84NGIPNGVLAMINMKK37 pKa = 9.84ARR39 pKa = 11.84CVINSEE45 pKa = 4.08KK46 pKa = 11.05VKK48 pKa = 10.85VNKK51 pKa = 10.45LCGKK55 pKa = 10.06SSLSKK60 pKa = 10.32GRR62 pKa = 11.84RR63 pKa = 11.84AARR66 pKa = 11.84LAICSFCYY74 pKa = 9.31RR75 pKa = 11.84TNCTSGYY82 pKa = 9.91RR83 pKa = 11.84CLKK86 pKa = 9.43SRR88 pKa = 11.84NGINARR94 pKa = 11.84FEE96 pKa = 3.7KK97 pKa = 10.93AEE99 pKa = 3.74WVKK102 pKa = 10.91YY103 pKa = 10.05GKK105 pKa = 10.16SSSLFEE111 pKa = 4.34SEE113 pKa = 4.62TPICAPSLRR122 pKa = 11.84NHH124 pKa = 6.88IEE126 pKa = 3.79DD127 pKa = 2.99EE128 pKa = 4.38MNRR131 pKa = 11.84VRR133 pKa = 11.84HH134 pKa = 5.11SS135 pKa = 3.08
MM1 pKa = 7.71SFNPKK6 pKa = 9.57NNKK9 pKa = 8.48DD10 pKa = 3.35DD11 pKa = 3.78RR12 pKa = 11.84RR13 pKa = 11.84IFSLCLSMMRR23 pKa = 11.84NGIPNGVLAMINMKK37 pKa = 9.84ARR39 pKa = 11.84CVINSEE45 pKa = 4.08KK46 pKa = 11.05VKK48 pKa = 10.85VNKK51 pKa = 10.45LCGKK55 pKa = 10.06SSLSKK60 pKa = 10.32GRR62 pKa = 11.84RR63 pKa = 11.84AARR66 pKa = 11.84LAICSFCYY74 pKa = 9.31RR75 pKa = 11.84TNCTSGYY82 pKa = 9.91RR83 pKa = 11.84CLKK86 pKa = 9.43SRR88 pKa = 11.84NGINARR94 pKa = 11.84FEE96 pKa = 3.7KK97 pKa = 10.93AEE99 pKa = 3.74WVKK102 pKa = 10.91YY103 pKa = 10.05GKK105 pKa = 10.16SSSLFEE111 pKa = 4.34SEE113 pKa = 4.62TPICAPSLRR122 pKa = 11.84NHH124 pKa = 6.88IEE126 pKa = 3.79DD127 pKa = 2.99EE128 pKa = 4.38MNRR131 pKa = 11.84VRR133 pKa = 11.84HH134 pKa = 5.11SS135 pKa = 3.08
Molecular weight: 15.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2637 |
135 |
1887 |
659.3 |
75.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.93 ± 0.762 | 2.427 ± 0.576 |
5.65 ± 1.022 | 6.902 ± 0.362 |
5.688 ± 0.472 | 6.333 ± 0.609 |
1.972 ± 0.159 | 6.03 ± 0.541 |
7.774 ± 0.72 | 9.063 ± 1.232 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.369 | 5.044 ± 1.332 |
3.413 ± 0.137 | 3.034 ± 0.698 |
6.447 ± 0.725 | 8.95 ± 1.286 |
4.361 ± 0.654 | 5.65 ± 0.928 |
0.872 ± 0.189 | 3.034 ± 0.347 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
