
Feline stool-associated circular virus KU14
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 7.64
Get precalculated fractions of proteins
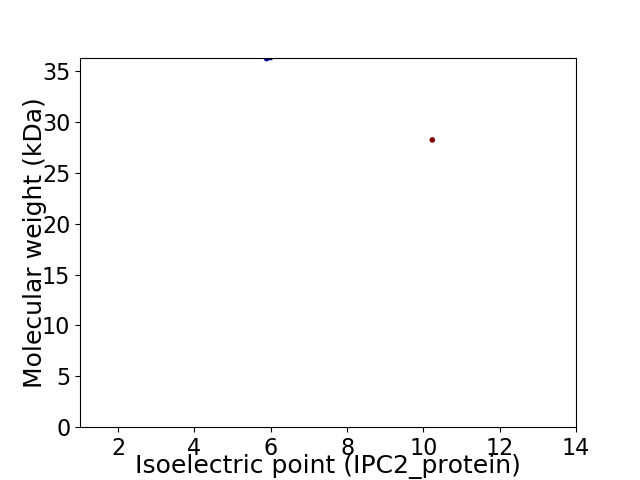
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
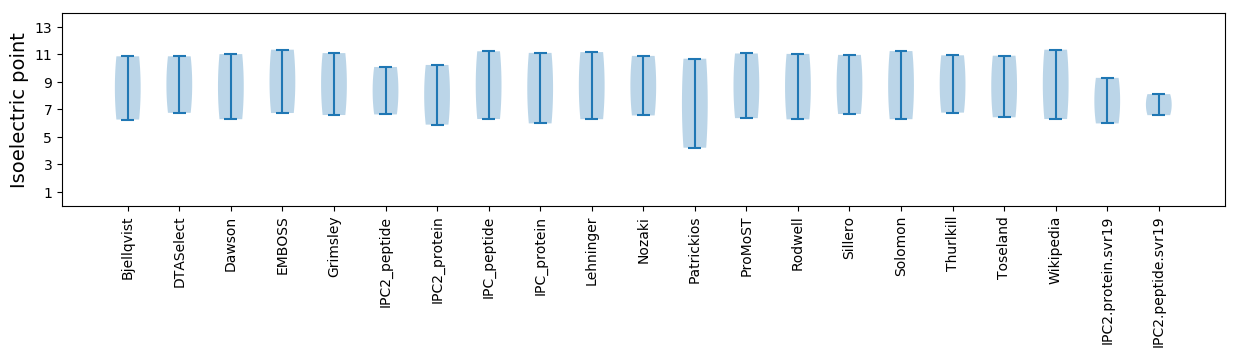
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
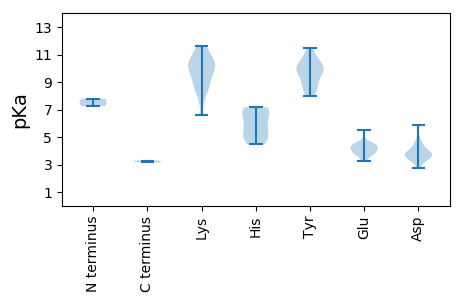
>tr|A0A2Z6GFU3|A0A2Z6GFU3_9CIRC ATP-dependent helicase Rep OS=Feline stool-associated circular virus KU14 OX=2249941 GN=putative Rep PE=3 SV=1
MM1 pKa = 7.79AATNWCGTLHH11 pKa = 7.04LGDD14 pKa = 3.84EE15 pKa = 4.42EE16 pKa = 5.52FYY18 pKa = 11.49GEE20 pKa = 3.95SWLNQLLGAGKK31 pKa = 8.03ITYY34 pKa = 10.12GIGQIEE40 pKa = 4.69SCPEE44 pKa = 3.26TGRR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.48FQFYY53 pKa = 9.04VQVGRR58 pKa = 11.84TQKK61 pKa = 9.56LAWMRR66 pKa = 11.84RR67 pKa = 11.84HH68 pKa = 6.4ISDD71 pKa = 3.66KK72 pKa = 11.28AHH74 pKa = 6.49FEE76 pKa = 3.9QARR79 pKa = 11.84GNVQQNIDD87 pKa = 3.76YY88 pKa = 10.56CSKK91 pKa = 10.43QDD93 pKa = 4.01SRR95 pKa = 11.84LDD97 pKa = 3.82GPWEE101 pKa = 3.72VGIARR106 pKa = 11.84RR107 pKa = 11.84QGQTTGLTAATEE119 pKa = 4.44MIASGAAVCEE129 pKa = 4.22VAKK132 pKa = 10.28LYY134 pKa = 9.99PVVWVRR140 pKa = 11.84HH141 pKa = 4.7GKK143 pKa = 10.29GLVHH147 pKa = 7.16LRR149 pKa = 11.84QTLALDD155 pKa = 3.65ADD157 pKa = 3.92RR158 pKa = 11.84RR159 pKa = 11.84EE160 pKa = 4.27FGPDD164 pKa = 3.55GPEE167 pKa = 3.42LWVLYY172 pKa = 10.28GPSGGGKK179 pKa = 9.15SRR181 pKa = 11.84FAKK184 pKa = 9.01EE185 pKa = 3.71HH186 pKa = 5.99WPDD189 pKa = 3.79AFWKK193 pKa = 10.53APYY196 pKa = 8.82SQWWDD201 pKa = 3.38GYY203 pKa = 9.87GKK205 pKa = 9.94HH206 pKa = 5.36EE207 pKa = 4.42TVILDD212 pKa = 3.72DD213 pKa = 4.85FKK215 pKa = 11.59DD216 pKa = 3.68DD217 pKa = 4.08SMRR220 pKa = 11.84LADD223 pKa = 4.51LQRR226 pKa = 11.84LIDD229 pKa = 4.69WYY231 pKa = 9.71PLWVEE236 pKa = 4.28VKK238 pKa = 10.55GGSLPMLATRR248 pKa = 11.84YY249 pKa = 9.99VITSNTHH256 pKa = 5.59PEE258 pKa = 3.69TWYY261 pKa = 9.6TRR263 pKa = 11.84ADD265 pKa = 3.26VHH267 pKa = 4.95KK268 pKa = 8.49TIWRR272 pKa = 11.84RR273 pKa = 11.84VQDD276 pKa = 3.52FAEE279 pKa = 3.95KK280 pKa = 10.14HH281 pKa = 5.05GRR283 pKa = 11.84LLRR286 pKa = 11.84FPPGEE291 pKa = 3.98AAAEE295 pKa = 3.91EE296 pKa = 4.47AAAEE300 pKa = 4.43PGSADD305 pKa = 3.38YY306 pKa = 11.38LGFPDD311 pKa = 5.91VFNSWNPLL319 pKa = 3.29
MM1 pKa = 7.79AATNWCGTLHH11 pKa = 7.04LGDD14 pKa = 3.84EE15 pKa = 4.42EE16 pKa = 5.52FYY18 pKa = 11.49GEE20 pKa = 3.95SWLNQLLGAGKK31 pKa = 8.03ITYY34 pKa = 10.12GIGQIEE40 pKa = 4.69SCPEE44 pKa = 3.26TGRR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.48FQFYY53 pKa = 9.04VQVGRR58 pKa = 11.84TQKK61 pKa = 9.56LAWMRR66 pKa = 11.84RR67 pKa = 11.84HH68 pKa = 6.4ISDD71 pKa = 3.66KK72 pKa = 11.28AHH74 pKa = 6.49FEE76 pKa = 3.9QARR79 pKa = 11.84GNVQQNIDD87 pKa = 3.76YY88 pKa = 10.56CSKK91 pKa = 10.43QDD93 pKa = 4.01SRR95 pKa = 11.84LDD97 pKa = 3.82GPWEE101 pKa = 3.72VGIARR106 pKa = 11.84RR107 pKa = 11.84QGQTTGLTAATEE119 pKa = 4.44MIASGAAVCEE129 pKa = 4.22VAKK132 pKa = 10.28LYY134 pKa = 9.99PVVWVRR140 pKa = 11.84HH141 pKa = 4.7GKK143 pKa = 10.29GLVHH147 pKa = 7.16LRR149 pKa = 11.84QTLALDD155 pKa = 3.65ADD157 pKa = 3.92RR158 pKa = 11.84RR159 pKa = 11.84EE160 pKa = 4.27FGPDD164 pKa = 3.55GPEE167 pKa = 3.42LWVLYY172 pKa = 10.28GPSGGGKK179 pKa = 9.15SRR181 pKa = 11.84FAKK184 pKa = 9.01EE185 pKa = 3.71HH186 pKa = 5.99WPDD189 pKa = 3.79AFWKK193 pKa = 10.53APYY196 pKa = 8.82SQWWDD201 pKa = 3.38GYY203 pKa = 9.87GKK205 pKa = 9.94HH206 pKa = 5.36EE207 pKa = 4.42TVILDD212 pKa = 3.72DD213 pKa = 4.85FKK215 pKa = 11.59DD216 pKa = 3.68DD217 pKa = 4.08SMRR220 pKa = 11.84LADD223 pKa = 4.51LQRR226 pKa = 11.84LIDD229 pKa = 4.69WYY231 pKa = 9.71PLWVEE236 pKa = 4.28VKK238 pKa = 10.55GGSLPMLATRR248 pKa = 11.84YY249 pKa = 9.99VITSNTHH256 pKa = 5.59PEE258 pKa = 3.69TWYY261 pKa = 9.6TRR263 pKa = 11.84ADD265 pKa = 3.26VHH267 pKa = 4.95KK268 pKa = 8.49TIWRR272 pKa = 11.84RR273 pKa = 11.84VQDD276 pKa = 3.52FAEE279 pKa = 3.95KK280 pKa = 10.14HH281 pKa = 5.05GRR283 pKa = 11.84LLRR286 pKa = 11.84FPPGEE291 pKa = 3.98AAAEE295 pKa = 3.91EE296 pKa = 4.47AAAEE300 pKa = 4.43PGSADD305 pKa = 3.38YY306 pKa = 11.38LGFPDD311 pKa = 5.91VFNSWNPLL319 pKa = 3.29
Molecular weight: 36.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z6GFU3|A0A2Z6GFU3_9CIRC ATP-dependent helicase Rep OS=Feline stool-associated circular virus KU14 OX=2249941 GN=putative Rep PE=3 SV=1
MM1 pKa = 7.26GMGRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 8.49YY9 pKa = 9.64RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84YY13 pKa = 8.81VSRR16 pKa = 11.84PPRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84MTYY24 pKa = 8.09RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 9.51SRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 8.0CRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84APKK39 pKa = 8.26GTYY42 pKa = 9.85SSVVTLTQDD51 pKa = 4.15AEE53 pKa = 4.49WKK55 pKa = 10.34FSNQNASNRR64 pKa = 11.84WVAFTFSPVSVPGFLDD80 pKa = 3.47YY81 pKa = 11.04KK82 pKa = 9.87ATYY85 pKa = 9.33SHH87 pKa = 7.01FRR89 pKa = 11.84ILKK92 pKa = 8.91AKK94 pKa = 10.21LYY96 pKa = 10.53LSRR99 pKa = 11.84ALGNDD104 pKa = 2.72AGTTFNYY111 pKa = 10.19LVVGSRR117 pKa = 11.84PFAATQNAGTTTGQDD132 pKa = 3.26ALVPAQTEE140 pKa = 3.94EE141 pKa = 4.27ALRR144 pKa = 11.84QAKK147 pKa = 6.6WQRR150 pKa = 11.84VRR152 pKa = 11.84YY153 pKa = 8.74PSTTTNVVPVGFHH166 pKa = 7.09PYY168 pKa = 10.48AIVQTFGPTPYY179 pKa = 8.24TTGSDD184 pKa = 3.71MMWQRR189 pKa = 11.84IWEE192 pKa = 4.25GRR194 pKa = 11.84RR195 pKa = 11.84WMPFSWAAATGSALSFFGPYY215 pKa = 10.25LVVDD219 pKa = 4.29KK220 pKa = 11.65SNGEE224 pKa = 3.96LSQGSWSVQCTLKK237 pKa = 10.65ISVQFKK243 pKa = 8.92GQRR246 pKa = 3.15
MM1 pKa = 7.26GMGRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 8.49YY9 pKa = 9.64RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84YY13 pKa = 8.81VSRR16 pKa = 11.84PPRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84MTYY24 pKa = 8.09RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 9.51SRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 8.0CRR33 pKa = 11.84TRR35 pKa = 11.84RR36 pKa = 11.84APKK39 pKa = 8.26GTYY42 pKa = 9.85SSVVTLTQDD51 pKa = 4.15AEE53 pKa = 4.49WKK55 pKa = 10.34FSNQNASNRR64 pKa = 11.84WVAFTFSPVSVPGFLDD80 pKa = 3.47YY81 pKa = 11.04KK82 pKa = 9.87ATYY85 pKa = 9.33SHH87 pKa = 7.01FRR89 pKa = 11.84ILKK92 pKa = 8.91AKK94 pKa = 10.21LYY96 pKa = 10.53LSRR99 pKa = 11.84ALGNDD104 pKa = 2.72AGTTFNYY111 pKa = 10.19LVVGSRR117 pKa = 11.84PFAATQNAGTTTGQDD132 pKa = 3.26ALVPAQTEE140 pKa = 3.94EE141 pKa = 4.27ALRR144 pKa = 11.84QAKK147 pKa = 6.6WQRR150 pKa = 11.84VRR152 pKa = 11.84YY153 pKa = 8.74PSTTTNVVPVGFHH166 pKa = 7.09PYY168 pKa = 10.48AIVQTFGPTPYY179 pKa = 8.24TTGSDD184 pKa = 3.71MMWQRR189 pKa = 11.84IWEE192 pKa = 4.25GRR194 pKa = 11.84RR195 pKa = 11.84WMPFSWAAATGSALSFFGPYY215 pKa = 10.25LVVDD219 pKa = 4.29KK220 pKa = 11.65SNGEE224 pKa = 3.96LSQGSWSVQCTLKK237 pKa = 10.65ISVQFKK243 pKa = 8.92GQRR246 pKa = 3.15
Molecular weight: 28.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
565 |
246 |
319 |
282.5 |
32.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.673 ± 0.358 | 1.062 ± 0.164 |
4.602 ± 1.429 | 4.425 ± 1.581 |
4.425 ± 0.568 | 8.673 ± 0.896 |
2.301 ± 0.983 | 2.655 ± 0.68 |
4.071 ± 0.272 | 6.903 ± 1.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.947 ± 0.325 | 2.655 ± 0.395 |
5.31 ± 0.252 | 4.956 ± 0.217 |
8.85 ± 1.673 | 6.195 ± 1.547 |
7.08 ± 1.5 | 6.372 ± 0.625 |
4.071 ± 0.541 | 4.779 ± 0.871 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
