
Janthinobacterium sp. HH01
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Janthinobacterium; unclassified Janthinobacterium
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
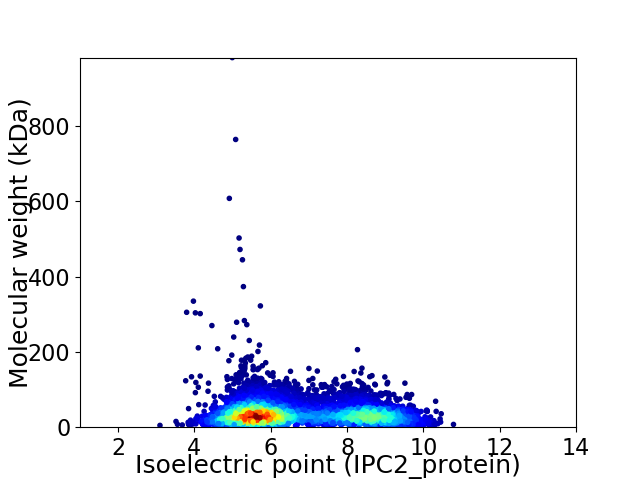
Virtual 2D-PAGE plot for 5974 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
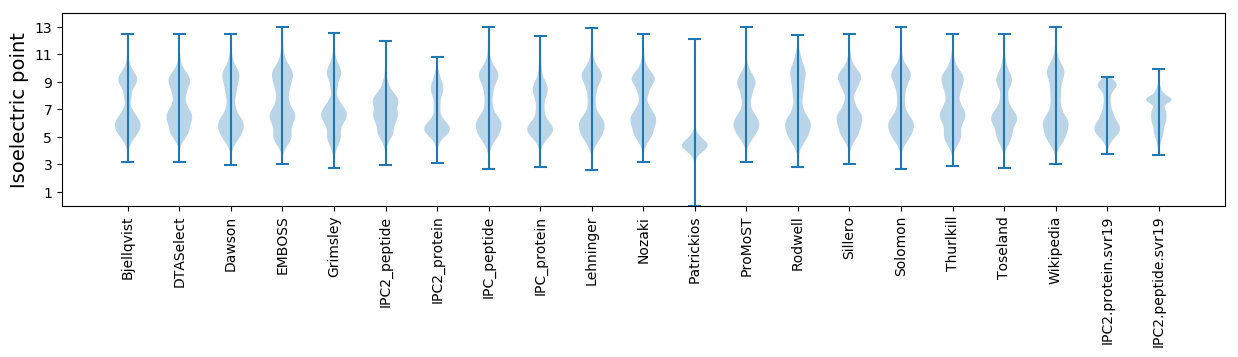
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L9PM13|L9PM13_9BURK Thiazole synthase OS=Janthinobacterium sp. HH01 OX=1198452 GN=thiG PE=3 SV=1
MM1 pKa = 6.34YY2 pKa = 7.89TALFTPAANVSGLASVTVTGGSYY25 pKa = 10.71TDD27 pKa = 3.95LLGNQGGAAVTPVIAVDD44 pKa = 3.57TRR46 pKa = 11.84PPTLAITSDD55 pKa = 3.66LAAVHH60 pKa = 6.59IGDD63 pKa = 3.82TATVSFTFSEE73 pKa = 4.68LPVGFAAGDD82 pKa = 3.33ITTTGGALSGFGVTANPLVYY102 pKa = 9.54TAIFTPAAGVASGAASISVAGGSYY126 pKa = 10.62FDD128 pKa = 4.4LAGNSGAAGATPSIAIDD145 pKa = 3.46TLAPTVTVGGVAFSADD161 pKa = 3.41TGASASDD168 pKa = 3.52LVTRR172 pKa = 11.84SAAQTVSGTLSAALLAGEE190 pKa = 4.49VVQVSLDD197 pKa = 4.04HH198 pKa = 5.91GQSWQNASGGAGSASWSLAGVTLAGSDD225 pKa = 3.61TLQVRR230 pKa = 11.84VNDD233 pKa = 3.63AAGNHH238 pKa = 5.71GAAFSAAYY246 pKa = 10.36VLDD249 pKa = 3.63ATAPTVLVASDD260 pKa = 4.6LGALNAGQQAHH271 pKa = 5.45LTFTFSEE278 pKa = 4.7APSGLTLASLSATGGAVSALAATANPLVYY307 pKa = 9.44TALFTPAAGQAGVSASVAVNAGGYY331 pKa = 8.27TDD333 pKa = 4.46LAGNGGQSGASLPIVINSALPSVLITSSAAALKK366 pKa = 10.66SGEE369 pKa = 4.11TALISFTFSTPPTGFTAADD388 pKa = 3.36IAVSAGTGALSGFAATANPLVYY410 pKa = 9.44TALFTPAAGQAAASAAVSVAGGSYY434 pKa = 10.67TDD436 pKa = 3.28VFGNNGGGGSSPSIAIDD453 pKa = 3.31TLAPTVSISSSVAGVKK469 pKa = 9.79IGEE472 pKa = 4.34TAVITFTFSEE482 pKa = 4.47QPLGFTAGDD491 pKa = 3.71VVTTGGALSNLAVSADD507 pKa = 3.66PLVYY511 pKa = 9.43TALFTPTAGLQSTAASVSIGAGAFVDD537 pKa = 4.29LAGNGATAGASPSLAVDD554 pKa = 3.75TLAPSAVANTVLFSADD570 pKa = 3.44SGVSATDD577 pKa = 3.36LVTRR581 pKa = 11.84TAAQTISGTLATALGAGEE599 pKa = 4.18VVEE602 pKa = 4.71VSLNNGASWSAATAGAGNSWTLAGQTLAGSNTLQVRR638 pKa = 11.84VSDD641 pKa = 3.54AAGNHH646 pKa = 5.46GAAFSTAYY654 pKa = 10.83VLDD657 pKa = 3.76NTAPTAAISSSAAALKK673 pKa = 10.67AGEE676 pKa = 4.26TATLTFTFSEE686 pKa = 4.68APSGLGAGSVTVSGGVLGAFTATANPLVYY715 pKa = 9.36TALFTPTAGVDD726 pKa = 3.3GGSAGIALNAGGYY739 pKa = 10.0SDD741 pKa = 3.7TAGNSGGGYY750 pKa = 6.57TLPAIAVDD758 pKa = 3.9TQAPSIAGASVALSADD774 pKa = 3.51SGVAADD780 pKa = 5.73LITNIAAQTLSGTLNGNLSAGDD802 pKa = 3.8TVEE805 pKa = 4.28VSLDD809 pKa = 3.49NGASWSAAAGAVGTAAWTLAGQTLTAGAGHH839 pKa = 5.41QVQVRR844 pKa = 11.84VSDD847 pKa = 3.39AAGNHH852 pKa = 5.76GAVASSAYY860 pKa = 10.69VLDD863 pKa = 3.93QSAPTLQISSSKK875 pKa = 9.55TVLNSAEE882 pKa = 4.28TATLTFTFSEE892 pKa = 4.73APQGFTAGAIAATGGAVSGFAATGGAVSGFAATANPLVYY931 pKa = 9.46TAVFTPTAGQAAGVGAVSVAAGAYY955 pKa = 8.26TDD957 pKa = 3.74AAGNSGLAGAGPAISYY973 pKa = 7.82ATVAPGVAISSDD985 pKa = 3.55VSALHH990 pKa = 6.86AGQTAIISFKK1000 pKa = 10.78FSVPPLGFTASDD1012 pKa = 3.33VSTSNGTLGALTATADD1028 pKa = 3.82PLLYY1032 pKa = 9.95TAVFTPAANLAGGGGGVSIGAGLFVDD1058 pKa = 4.76GLGNAGSAAAMAPISIDD1075 pKa = 3.18TLAPTLAITSGAAALNSGATSLITFTFSEE1104 pKa = 4.65APPAFSLGDD1113 pKa = 3.18ITVANGTLANLVQSANPLVYY1133 pKa = 9.48TAVFTPGANIAAGAATIAVIGGAYY1157 pKa = 8.73TDD1159 pKa = 3.86LAGNGGGAAPTPSISIDD1176 pKa = 3.27TLAPTIAGASLLFSADD1192 pKa = 3.35TGSSNTDD1199 pKa = 4.19LITRR1203 pKa = 11.84TAAQTIAGTVNGLLAAGDD1221 pKa = 3.9VVEE1224 pKa = 4.67VSFDD1228 pKa = 3.63SGASWVGASAAAGGSSWVLAGQTLGGSGTLQVRR1261 pKa = 11.84VSDD1264 pKa = 3.86ANGNHH1269 pKa = 5.87GAVYY1273 pKa = 10.46ARR1275 pKa = 11.84AYY1277 pKa = 10.79VLDD1280 pKa = 3.87TTAPTVAISSDD1291 pKa = 3.2TATLRR1296 pKa = 11.84NGQAATISFTFSEE1309 pKa = 4.83APAGFTLGDD1318 pKa = 4.54LIATGGTLSGFAATANPLVYY1338 pKa = 9.71TVLLTPTPGLAGAAGVTLGNGLYY1361 pKa = 10.54ADD1363 pKa = 3.98AAGNSGSGAGSPAISVDD1380 pKa = 3.99TIAPTLAIAASSAALKK1396 pKa = 10.93AGDD1399 pKa = 3.58TAVITFTFSDD1409 pKa = 3.83APAAFTLADD1418 pKa = 3.48VSASHH1423 pKa = 6.21GTLSALAAGPDD1434 pKa = 3.58PRR1436 pKa = 11.84VYY1438 pKa = 10.06TAVFTPDD1445 pKa = 3.25AGTAAASAVVSVAGSVYY1462 pKa = 11.04ADD1464 pKa = 3.18AAGNTGSSVSSGAIAIDD1481 pKa = 3.44TLAPTTSGATVVFSADD1497 pKa = 3.4YY1498 pKa = 9.98GASGSDD1504 pKa = 3.19LVTNAVGQVISGTLDD1519 pKa = 3.07APLAAGEE1526 pKa = 4.07QVQVSLDD1533 pKa = 3.73NGASWLNAVAAAGLSSWALAPQLLNGSGTLQVRR1566 pKa = 11.84VADD1569 pKa = 3.67AAGNHH1574 pKa = 5.87GPAFSAGYY1582 pKa = 10.32VLDD1585 pKa = 4.33LVAPAMTVTSSAAALKK1601 pKa = 10.71AGDD1604 pKa = 4.03SATLTFTFSEE1614 pKa = 4.7APTGFTQADD1623 pKa = 3.93LVALNGTLSGFTPTANPLVYY1643 pKa = 9.83TVVLTPAAGLGGVSAGVTLAAGLYY1667 pKa = 9.4TDD1669 pKa = 3.91VAGNPGAAAASPLVAIDD1686 pKa = 3.7TQPPTVAITTSATQLKK1702 pKa = 9.63IGEE1705 pKa = 4.52LATITFTFNEE1715 pKa = 4.26APLGFTAGDD1724 pKa = 3.64VAVTGGSLSGFGLTANPLVYY1744 pKa = 8.02TTLFTPTPGVAGGTASIAIGAGDD1767 pKa = 4.0YY1768 pKa = 11.12LDD1770 pKa = 4.93AAGNPGLAAAAPAIQFSTLAPATGGASVSFSADD1803 pKa = 2.37TGVFNNDD1810 pKa = 3.31LVTSSALQTINGTLDD1825 pKa = 3.0ANLAVGEE1832 pKa = 4.1AVEE1835 pKa = 4.3VSLDD1839 pKa = 3.66NGASWSTALAATGSPAWSLGPLVLGGSGVLQVRR1872 pKa = 11.84VTDD1875 pKa = 3.55AAGNHH1880 pKa = 5.75GAASASAYY1888 pKa = 10.75VLDD1891 pKa = 4.05TGAPGVAISADD1902 pKa = 3.44AAALKK1907 pKa = 10.76AGDD1910 pKa = 3.99SATLSFTFTEE1920 pKa = 4.8APHH1923 pKa = 6.7GFTTADD1929 pKa = 3.28VVAVGGIKK1937 pKa = 10.41GALTATANPLVYY1949 pKa = 10.09TMSYY1953 pKa = 9.3TPTAGFSGVGGVTLTAASYY1972 pKa = 10.77TDD1974 pKa = 3.92LAGNPGAGAAGPALSVDD1991 pKa = 3.78AQPPTLSITSSSTALKK2007 pKa = 10.68AGDD2010 pKa = 3.87SATLTFTFSEE2020 pKa = 4.63APLGFAAGDD2029 pKa = 3.51ITVSGGTIGGFSATANPLVYY2049 pKa = 9.46TAVFTPTPGTAAGSAVIAVASGVYY2073 pKa = 10.18SDD2075 pKa = 4.12AAGNGGASGSAPAIVIDD2092 pKa = 3.96TLAPLAAVGGAIAFSTDD2109 pKa = 2.86TGISNNDD2116 pKa = 4.09LITSVAAQTVSGTLSAPLAAGEE2138 pKa = 4.26TVQVSFDD2145 pKa = 4.38NGASWQAAAVSGTGWSAGVTLTGSGVMRR2173 pKa = 11.84LYY2175 pKa = 11.23VSDD2178 pKa = 3.56AAGNHH2183 pKa = 5.49SPEE2186 pKa = 3.85IVQAYY2191 pKa = 9.73QYY2193 pKa = 10.87DD2194 pKa = 3.91ATPPTVAITASAAVANGMAPTTITFSFSEE2223 pKa = 4.42APQGFTAADD2232 pKa = 3.71LTVSGGTLGAVTATANPLVYY2252 pKa = 9.61TVQFTPTPGQASGIATITLTGGGYY2276 pKa = 10.14VDD2278 pKa = 3.89AAGNNGAGNALPALSFDD2295 pKa = 3.97TMPPAAAAAGAQFSSDD2311 pKa = 3.19SGTPGDD2317 pKa = 4.65MITNNPSQTLTGTLSAPLAAGDD2339 pKa = 3.99VVQVSLDD2346 pKa = 4.13DD2347 pKa = 4.06GVNWNIATVSGTTWTLAGRR2366 pKa = 11.84TLQGSSRR2373 pKa = 11.84MVVRR2377 pKa = 11.84VSDD2380 pKa = 3.49AAGNVSTPMVMPYY2393 pKa = 10.69VLDD2396 pKa = 3.79TQAPTVTITSSVTQLGEE2413 pKa = 4.14GGSAQITLTLSDD2425 pKa = 4.4PGVLTLADD2433 pKa = 3.41IGVSGGTLSNLSGGGTVYY2451 pKa = 10.62TVMLTPPPFSVTPITVSVAGHH2472 pKa = 5.74VFSDD2476 pKa = 3.48AAGNASAAAAPLQLGVNTFPTPISGTPSTVDD2507 pKa = 3.06GVQIMTQTGIDD2518 pKa = 3.48ARR2520 pKa = 11.84TGVATRR2526 pKa = 11.84TVTVPIISDD2535 pKa = 3.51TRR2537 pKa = 11.84SEE2539 pKa = 4.18DD2540 pKa = 2.99HH2541 pKa = 6.3STPHH2545 pKa = 6.88ANLADD2550 pKa = 3.68IPLGLAAAGATPGTSLVVSLPVGVGLEE2577 pKa = 3.94AAGPSVLLGGATALTDD2593 pKa = 4.62LIGRR2597 pKa = 11.84IDD2599 pKa = 3.78DD2600 pKa = 3.84HH2601 pKa = 6.14TLQGQATRR2609 pKa = 11.84AAMEE2613 pKa = 4.35AQASAYY2619 pKa = 9.67LASLAPGVQLQQGTLTLGAASAVADD2644 pKa = 4.08GAEE2647 pKa = 4.0IMITGAGATAGGAPHH2662 pKa = 6.96GALYY2666 pKa = 8.43DD2667 pKa = 3.77TATGGTAVALVIDD2680 pKa = 4.9ARR2682 pKa = 11.84ALPQGIGLEE2691 pKa = 3.96MAGIDD2696 pKa = 3.41FAAFIGAATVRR2707 pKa = 11.84GGAGQNFFIGDD2718 pKa = 3.74AAAQRR2723 pKa = 11.84IVLTDD2728 pKa = 3.23GGGNDD2733 pKa = 3.45TLYY2736 pKa = 11.32GNGGNDD2742 pKa = 3.42VLGTAGGRR2750 pKa = 11.84DD2751 pKa = 4.02YY2752 pKa = 11.73LDD2754 pKa = 3.91GGAGNDD2760 pKa = 3.48ILFGGAGNDD2769 pKa = 3.74VLNGGADD2776 pKa = 3.57NDD2778 pKa = 4.29TLQGGRR2784 pKa = 11.84SDD2786 pKa = 3.36TGQWQFYY2793 pKa = 9.25LNPAGAVVGRR2803 pKa = 11.84HH2804 pKa = 5.04EE2805 pKa = 4.39MALTDD2810 pKa = 3.76AAATEE2815 pKa = 4.61TVTAAQLNQDD2825 pKa = 3.41VALLGFAGPKK2835 pKa = 9.37AANLQDD2841 pKa = 3.3WSLMYY2846 pKa = 10.23HH2847 pKa = 6.69AAYY2850 pKa = 9.91HH2851 pKa = 6.37RR2852 pKa = 11.84APDD2855 pKa = 4.34LGGLSYY2861 pKa = 10.53WSTSGGTPQQHH2872 pKa = 6.26AAGFLQSPEE2881 pKa = 3.88GLNGIMALNNHH2892 pKa = 6.54DD2893 pKa = 4.61FVVQLLQNSVGRR2905 pKa = 11.84AGTAAEE2911 pKa = 4.26LSRR2914 pKa = 11.84WIGMLDD2920 pKa = 3.59AAPGDD2925 pKa = 3.84MQVRR2929 pKa = 11.84GAVFTDD2935 pKa = 3.26IALSAEE2941 pKa = 3.88HH2942 pKa = 6.56RR2943 pKa = 11.84AGYY2946 pKa = 7.96ITADD2950 pKa = 3.64GVPLGGEE2957 pKa = 4.06LLADD2961 pKa = 3.56EE2962 pKa = 5.47RR2963 pKa = 11.84GWIAGSGDD2971 pKa = 3.65DD2972 pKa = 4.14RR2973 pKa = 11.84LQGGGGNDD2981 pKa = 3.83LLVGGDD2987 pKa = 3.71GVDD2990 pKa = 3.28TVVYY2994 pKa = 10.28GDD2996 pKa = 3.89AASSHH3001 pKa = 5.49SVVLGRR3007 pKa = 11.84TGEE3010 pKa = 4.14VMIAAPDD3017 pKa = 3.66GGRR3020 pKa = 11.84DD3021 pKa = 3.33TLRR3024 pKa = 11.84QIEE3027 pKa = 4.11RR3028 pKa = 11.84GEE3030 pKa = 4.1FKK3032 pKa = 10.87GGEE3035 pKa = 4.24TVDD3038 pKa = 4.69LGFTQASAKK3047 pKa = 10.19ALQEE3051 pKa = 3.95LGMMYY3056 pKa = 10.53HH3057 pKa = 6.34LTLNRR3062 pKa = 11.84AADD3065 pKa = 3.72LGGFQFWLASGLEE3078 pKa = 4.38GSALAHH3084 pKa = 6.55GFTDD3088 pKa = 3.56STEE3091 pKa = 3.69MAQRR3095 pKa = 11.84FGQLDD3100 pKa = 3.37DD3101 pKa = 4.29AGFVKK3106 pKa = 10.77LLYY3109 pKa = 10.44QNTVRR3114 pKa = 11.84HH3115 pKa = 6.41DD3116 pKa = 3.49PDD3118 pKa = 3.6AATLARR3124 pKa = 11.84WDD3126 pKa = 3.82SFLDD3130 pKa = 3.36SHH3132 pKa = 6.94HH3133 pKa = 6.77RR3134 pKa = 11.84ADD3136 pKa = 5.46LVALLASDD3144 pKa = 3.55VTLIGSQYY3152 pKa = 9.46GTDD3155 pKa = 3.32GMNLIGGLL3163 pKa = 3.73
MM1 pKa = 6.34YY2 pKa = 7.89TALFTPAANVSGLASVTVTGGSYY25 pKa = 10.71TDD27 pKa = 3.95LLGNQGGAAVTPVIAVDD44 pKa = 3.57TRR46 pKa = 11.84PPTLAITSDD55 pKa = 3.66LAAVHH60 pKa = 6.59IGDD63 pKa = 3.82TATVSFTFSEE73 pKa = 4.68LPVGFAAGDD82 pKa = 3.33ITTTGGALSGFGVTANPLVYY102 pKa = 9.54TAIFTPAAGVASGAASISVAGGSYY126 pKa = 10.62FDD128 pKa = 4.4LAGNSGAAGATPSIAIDD145 pKa = 3.46TLAPTVTVGGVAFSADD161 pKa = 3.41TGASASDD168 pKa = 3.52LVTRR172 pKa = 11.84SAAQTVSGTLSAALLAGEE190 pKa = 4.49VVQVSLDD197 pKa = 4.04HH198 pKa = 5.91GQSWQNASGGAGSASWSLAGVTLAGSDD225 pKa = 3.61TLQVRR230 pKa = 11.84VNDD233 pKa = 3.63AAGNHH238 pKa = 5.71GAAFSAAYY246 pKa = 10.36VLDD249 pKa = 3.63ATAPTVLVASDD260 pKa = 4.6LGALNAGQQAHH271 pKa = 5.45LTFTFSEE278 pKa = 4.7APSGLTLASLSATGGAVSALAATANPLVYY307 pKa = 9.44TALFTPAAGQAGVSASVAVNAGGYY331 pKa = 8.27TDD333 pKa = 4.46LAGNGGQSGASLPIVINSALPSVLITSSAAALKK366 pKa = 10.66SGEE369 pKa = 4.11TALISFTFSTPPTGFTAADD388 pKa = 3.36IAVSAGTGALSGFAATANPLVYY410 pKa = 9.44TALFTPAAGQAAASAAVSVAGGSYY434 pKa = 10.67TDD436 pKa = 3.28VFGNNGGGGSSPSIAIDD453 pKa = 3.31TLAPTVSISSSVAGVKK469 pKa = 9.79IGEE472 pKa = 4.34TAVITFTFSEE482 pKa = 4.47QPLGFTAGDD491 pKa = 3.71VVTTGGALSNLAVSADD507 pKa = 3.66PLVYY511 pKa = 9.43TALFTPTAGLQSTAASVSIGAGAFVDD537 pKa = 4.29LAGNGATAGASPSLAVDD554 pKa = 3.75TLAPSAVANTVLFSADD570 pKa = 3.44SGVSATDD577 pKa = 3.36LVTRR581 pKa = 11.84TAAQTISGTLATALGAGEE599 pKa = 4.18VVEE602 pKa = 4.71VSLNNGASWSAATAGAGNSWTLAGQTLAGSNTLQVRR638 pKa = 11.84VSDD641 pKa = 3.54AAGNHH646 pKa = 5.46GAAFSTAYY654 pKa = 10.83VLDD657 pKa = 3.76NTAPTAAISSSAAALKK673 pKa = 10.67AGEE676 pKa = 4.26TATLTFTFSEE686 pKa = 4.68APSGLGAGSVTVSGGVLGAFTATANPLVYY715 pKa = 9.36TALFTPTAGVDD726 pKa = 3.3GGSAGIALNAGGYY739 pKa = 10.0SDD741 pKa = 3.7TAGNSGGGYY750 pKa = 6.57TLPAIAVDD758 pKa = 3.9TQAPSIAGASVALSADD774 pKa = 3.51SGVAADD780 pKa = 5.73LITNIAAQTLSGTLNGNLSAGDD802 pKa = 3.8TVEE805 pKa = 4.28VSLDD809 pKa = 3.49NGASWSAAAGAVGTAAWTLAGQTLTAGAGHH839 pKa = 5.41QVQVRR844 pKa = 11.84VSDD847 pKa = 3.39AAGNHH852 pKa = 5.76GAVASSAYY860 pKa = 10.69VLDD863 pKa = 3.93QSAPTLQISSSKK875 pKa = 9.55TVLNSAEE882 pKa = 4.28TATLTFTFSEE892 pKa = 4.73APQGFTAGAIAATGGAVSGFAATGGAVSGFAATANPLVYY931 pKa = 9.46TAVFTPTAGQAAGVGAVSVAAGAYY955 pKa = 8.26TDD957 pKa = 3.74AAGNSGLAGAGPAISYY973 pKa = 7.82ATVAPGVAISSDD985 pKa = 3.55VSALHH990 pKa = 6.86AGQTAIISFKK1000 pKa = 10.78FSVPPLGFTASDD1012 pKa = 3.33VSTSNGTLGALTATADD1028 pKa = 3.82PLLYY1032 pKa = 9.95TAVFTPAANLAGGGGGVSIGAGLFVDD1058 pKa = 4.76GLGNAGSAAAMAPISIDD1075 pKa = 3.18TLAPTLAITSGAAALNSGATSLITFTFSEE1104 pKa = 4.65APPAFSLGDD1113 pKa = 3.18ITVANGTLANLVQSANPLVYY1133 pKa = 9.48TAVFTPGANIAAGAATIAVIGGAYY1157 pKa = 8.73TDD1159 pKa = 3.86LAGNGGGAAPTPSISIDD1176 pKa = 3.27TLAPTIAGASLLFSADD1192 pKa = 3.35TGSSNTDD1199 pKa = 4.19LITRR1203 pKa = 11.84TAAQTIAGTVNGLLAAGDD1221 pKa = 3.9VVEE1224 pKa = 4.67VSFDD1228 pKa = 3.63SGASWVGASAAAGGSSWVLAGQTLGGSGTLQVRR1261 pKa = 11.84VSDD1264 pKa = 3.86ANGNHH1269 pKa = 5.87GAVYY1273 pKa = 10.46ARR1275 pKa = 11.84AYY1277 pKa = 10.79VLDD1280 pKa = 3.87TTAPTVAISSDD1291 pKa = 3.2TATLRR1296 pKa = 11.84NGQAATISFTFSEE1309 pKa = 4.83APAGFTLGDD1318 pKa = 4.54LIATGGTLSGFAATANPLVYY1338 pKa = 9.71TVLLTPTPGLAGAAGVTLGNGLYY1361 pKa = 10.54ADD1363 pKa = 3.98AAGNSGSGAGSPAISVDD1380 pKa = 3.99TIAPTLAIAASSAALKK1396 pKa = 10.93AGDD1399 pKa = 3.58TAVITFTFSDD1409 pKa = 3.83APAAFTLADD1418 pKa = 3.48VSASHH1423 pKa = 6.21GTLSALAAGPDD1434 pKa = 3.58PRR1436 pKa = 11.84VYY1438 pKa = 10.06TAVFTPDD1445 pKa = 3.25AGTAAASAVVSVAGSVYY1462 pKa = 11.04ADD1464 pKa = 3.18AAGNTGSSVSSGAIAIDD1481 pKa = 3.44TLAPTTSGATVVFSADD1497 pKa = 3.4YY1498 pKa = 9.98GASGSDD1504 pKa = 3.19LVTNAVGQVISGTLDD1519 pKa = 3.07APLAAGEE1526 pKa = 4.07QVQVSLDD1533 pKa = 3.73NGASWLNAVAAAGLSSWALAPQLLNGSGTLQVRR1566 pKa = 11.84VADD1569 pKa = 3.67AAGNHH1574 pKa = 5.87GPAFSAGYY1582 pKa = 10.32VLDD1585 pKa = 4.33LVAPAMTVTSSAAALKK1601 pKa = 10.71AGDD1604 pKa = 4.03SATLTFTFSEE1614 pKa = 4.7APTGFTQADD1623 pKa = 3.93LVALNGTLSGFTPTANPLVYY1643 pKa = 9.83TVVLTPAAGLGGVSAGVTLAAGLYY1667 pKa = 9.4TDD1669 pKa = 3.91VAGNPGAAAASPLVAIDD1686 pKa = 3.7TQPPTVAITTSATQLKK1702 pKa = 9.63IGEE1705 pKa = 4.52LATITFTFNEE1715 pKa = 4.26APLGFTAGDD1724 pKa = 3.64VAVTGGSLSGFGLTANPLVYY1744 pKa = 8.02TTLFTPTPGVAGGTASIAIGAGDD1767 pKa = 4.0YY1768 pKa = 11.12LDD1770 pKa = 4.93AAGNPGLAAAAPAIQFSTLAPATGGASVSFSADD1803 pKa = 2.37TGVFNNDD1810 pKa = 3.31LVTSSALQTINGTLDD1825 pKa = 3.0ANLAVGEE1832 pKa = 4.1AVEE1835 pKa = 4.3VSLDD1839 pKa = 3.66NGASWSTALAATGSPAWSLGPLVLGGSGVLQVRR1872 pKa = 11.84VTDD1875 pKa = 3.55AAGNHH1880 pKa = 5.75GAASASAYY1888 pKa = 10.75VLDD1891 pKa = 4.05TGAPGVAISADD1902 pKa = 3.44AAALKK1907 pKa = 10.76AGDD1910 pKa = 3.99SATLSFTFTEE1920 pKa = 4.8APHH1923 pKa = 6.7GFTTADD1929 pKa = 3.28VVAVGGIKK1937 pKa = 10.41GALTATANPLVYY1949 pKa = 10.09TMSYY1953 pKa = 9.3TPTAGFSGVGGVTLTAASYY1972 pKa = 10.77TDD1974 pKa = 3.92LAGNPGAGAAGPALSVDD1991 pKa = 3.78AQPPTLSITSSSTALKK2007 pKa = 10.68AGDD2010 pKa = 3.87SATLTFTFSEE2020 pKa = 4.63APLGFAAGDD2029 pKa = 3.51ITVSGGTIGGFSATANPLVYY2049 pKa = 9.46TAVFTPTPGTAAGSAVIAVASGVYY2073 pKa = 10.18SDD2075 pKa = 4.12AAGNGGASGSAPAIVIDD2092 pKa = 3.96TLAPLAAVGGAIAFSTDD2109 pKa = 2.86TGISNNDD2116 pKa = 4.09LITSVAAQTVSGTLSAPLAAGEE2138 pKa = 4.26TVQVSFDD2145 pKa = 4.38NGASWQAAAVSGTGWSAGVTLTGSGVMRR2173 pKa = 11.84LYY2175 pKa = 11.23VSDD2178 pKa = 3.56AAGNHH2183 pKa = 5.49SPEE2186 pKa = 3.85IVQAYY2191 pKa = 9.73QYY2193 pKa = 10.87DD2194 pKa = 3.91ATPPTVAITASAAVANGMAPTTITFSFSEE2223 pKa = 4.42APQGFTAADD2232 pKa = 3.71LTVSGGTLGAVTATANPLVYY2252 pKa = 9.61TVQFTPTPGQASGIATITLTGGGYY2276 pKa = 10.14VDD2278 pKa = 3.89AAGNNGAGNALPALSFDD2295 pKa = 3.97TMPPAAAAAGAQFSSDD2311 pKa = 3.19SGTPGDD2317 pKa = 4.65MITNNPSQTLTGTLSAPLAAGDD2339 pKa = 3.99VVQVSLDD2346 pKa = 4.13DD2347 pKa = 4.06GVNWNIATVSGTTWTLAGRR2366 pKa = 11.84TLQGSSRR2373 pKa = 11.84MVVRR2377 pKa = 11.84VSDD2380 pKa = 3.49AAGNVSTPMVMPYY2393 pKa = 10.69VLDD2396 pKa = 3.79TQAPTVTITSSVTQLGEE2413 pKa = 4.14GGSAQITLTLSDD2425 pKa = 4.4PGVLTLADD2433 pKa = 3.41IGVSGGTLSNLSGGGTVYY2451 pKa = 10.62TVMLTPPPFSVTPITVSVAGHH2472 pKa = 5.74VFSDD2476 pKa = 3.48AAGNASAAAAPLQLGVNTFPTPISGTPSTVDD2507 pKa = 3.06GVQIMTQTGIDD2518 pKa = 3.48ARR2520 pKa = 11.84TGVATRR2526 pKa = 11.84TVTVPIISDD2535 pKa = 3.51TRR2537 pKa = 11.84SEE2539 pKa = 4.18DD2540 pKa = 2.99HH2541 pKa = 6.3STPHH2545 pKa = 6.88ANLADD2550 pKa = 3.68IPLGLAAAGATPGTSLVVSLPVGVGLEE2577 pKa = 3.94AAGPSVLLGGATALTDD2593 pKa = 4.62LIGRR2597 pKa = 11.84IDD2599 pKa = 3.78DD2600 pKa = 3.84HH2601 pKa = 6.14TLQGQATRR2609 pKa = 11.84AAMEE2613 pKa = 4.35AQASAYY2619 pKa = 9.67LASLAPGVQLQQGTLTLGAASAVADD2644 pKa = 4.08GAEE2647 pKa = 4.0IMITGAGATAGGAPHH2662 pKa = 6.96GALYY2666 pKa = 8.43DD2667 pKa = 3.77TATGGTAVALVIDD2680 pKa = 4.9ARR2682 pKa = 11.84ALPQGIGLEE2691 pKa = 3.96MAGIDD2696 pKa = 3.41FAAFIGAATVRR2707 pKa = 11.84GGAGQNFFIGDD2718 pKa = 3.74AAAQRR2723 pKa = 11.84IVLTDD2728 pKa = 3.23GGGNDD2733 pKa = 3.45TLYY2736 pKa = 11.32GNGGNDD2742 pKa = 3.42VLGTAGGRR2750 pKa = 11.84DD2751 pKa = 4.02YY2752 pKa = 11.73LDD2754 pKa = 3.91GGAGNDD2760 pKa = 3.48ILFGGAGNDD2769 pKa = 3.74VLNGGADD2776 pKa = 3.57NDD2778 pKa = 4.29TLQGGRR2784 pKa = 11.84SDD2786 pKa = 3.36TGQWQFYY2793 pKa = 9.25LNPAGAVVGRR2803 pKa = 11.84HH2804 pKa = 5.04EE2805 pKa = 4.39MALTDD2810 pKa = 3.76AAATEE2815 pKa = 4.61TVTAAQLNQDD2825 pKa = 3.41VALLGFAGPKK2835 pKa = 9.37AANLQDD2841 pKa = 3.3WSLMYY2846 pKa = 10.23HH2847 pKa = 6.69AAYY2850 pKa = 9.91HH2851 pKa = 6.37RR2852 pKa = 11.84APDD2855 pKa = 4.34LGGLSYY2861 pKa = 10.53WSTSGGTPQQHH2872 pKa = 6.26AAGFLQSPEE2881 pKa = 3.88GLNGIMALNNHH2892 pKa = 6.54DD2893 pKa = 4.61FVVQLLQNSVGRR2905 pKa = 11.84AGTAAEE2911 pKa = 4.26LSRR2914 pKa = 11.84WIGMLDD2920 pKa = 3.59AAPGDD2925 pKa = 3.84MQVRR2929 pKa = 11.84GAVFTDD2935 pKa = 3.26IALSAEE2941 pKa = 3.88HH2942 pKa = 6.56RR2943 pKa = 11.84AGYY2946 pKa = 7.96ITADD2950 pKa = 3.64GVPLGGEE2957 pKa = 4.06LLADD2961 pKa = 3.56EE2962 pKa = 5.47RR2963 pKa = 11.84GWIAGSGDD2971 pKa = 3.65DD2972 pKa = 4.14RR2973 pKa = 11.84LQGGGGNDD2981 pKa = 3.83LLVGGDD2987 pKa = 3.71GVDD2990 pKa = 3.28TVVYY2994 pKa = 10.28GDD2996 pKa = 3.89AASSHH3001 pKa = 5.49SVVLGRR3007 pKa = 11.84TGEE3010 pKa = 4.14VMIAAPDD3017 pKa = 3.66GGRR3020 pKa = 11.84DD3021 pKa = 3.33TLRR3024 pKa = 11.84QIEE3027 pKa = 4.11RR3028 pKa = 11.84GEE3030 pKa = 4.1FKK3032 pKa = 10.87GGEE3035 pKa = 4.24TVDD3038 pKa = 4.69LGFTQASAKK3047 pKa = 10.19ALQEE3051 pKa = 3.95LGMMYY3056 pKa = 10.53HH3057 pKa = 6.34LTLNRR3062 pKa = 11.84AADD3065 pKa = 3.72LGGFQFWLASGLEE3078 pKa = 4.38GSALAHH3084 pKa = 6.55GFTDD3088 pKa = 3.56STEE3091 pKa = 3.69MAQRR3095 pKa = 11.84FGQLDD3100 pKa = 3.37DD3101 pKa = 4.29AGFVKK3106 pKa = 10.77LLYY3109 pKa = 10.44QNTVRR3114 pKa = 11.84HH3115 pKa = 6.41DD3116 pKa = 3.49PDD3118 pKa = 3.6AATLARR3124 pKa = 11.84WDD3126 pKa = 3.82SFLDD3130 pKa = 3.36SHH3132 pKa = 6.94HH3133 pKa = 6.77RR3134 pKa = 11.84ADD3136 pKa = 5.46LVALLASDD3144 pKa = 3.55VTLIGSQYY3152 pKa = 9.46GTDD3155 pKa = 3.32GMNLIGGLL3163 pKa = 3.73
Molecular weight: 305.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L9PC82|L9PC82_9BURK Flagellar hook-length control protein OS=Janthinobacterium sp. HH01 OX=1198452 GN=Jab_2c12740 PE=4 SV=1
MM1 pKa = 7.91PKK3 pKa = 9.63MKK5 pKa = 9.83TKK7 pKa = 10.72SSAKK11 pKa = 9.15KK12 pKa = 9.67RR13 pKa = 11.84FRR15 pKa = 11.84VRR17 pKa = 11.84PGGTVKK23 pKa = 10.54SGMAFKK29 pKa = 10.69RR30 pKa = 11.84HH31 pKa = 5.84ILTKK35 pKa = 9.78KK36 pKa = 3.98TTKK39 pKa = 10.23NKK41 pKa = 9.21RR42 pKa = 11.84QLRR45 pKa = 11.84GTRR48 pKa = 11.84NINASDD54 pKa = 3.65VTSVMRR60 pKa = 11.84MMPTAA65 pKa = 4.09
MM1 pKa = 7.91PKK3 pKa = 9.63MKK5 pKa = 9.83TKK7 pKa = 10.72SSAKK11 pKa = 9.15KK12 pKa = 9.67RR13 pKa = 11.84FRR15 pKa = 11.84VRR17 pKa = 11.84PGGTVKK23 pKa = 10.54SGMAFKK29 pKa = 10.69RR30 pKa = 11.84HH31 pKa = 5.84ILTKK35 pKa = 9.78KK36 pKa = 3.98TTKK39 pKa = 10.23NKK41 pKa = 9.21RR42 pKa = 11.84QLRR45 pKa = 11.84GTRR48 pKa = 11.84NINASDD54 pKa = 3.65VTSVMRR60 pKa = 11.84MMPTAA65 pKa = 4.09
Molecular weight: 7.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2157853 |
29 |
9114 |
361.2 |
39.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.79 ± 0.054 | 0.835 ± 0.01 |
5.393 ± 0.02 | 5.012 ± 0.029 |
3.489 ± 0.021 | 8.073 ± 0.031 |
2.166 ± 0.015 | 4.659 ± 0.022 |
3.665 ± 0.031 | 10.684 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.561 ± 0.019 | 3.123 ± 0.023 |
4.891 ± 0.025 | 4.227 ± 0.028 |
6.158 ± 0.029 | 5.784 ± 0.025 |
5.307 ± 0.037 | 7.207 ± 0.028 |
1.346 ± 0.012 | 2.631 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
