
Beihai narna-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
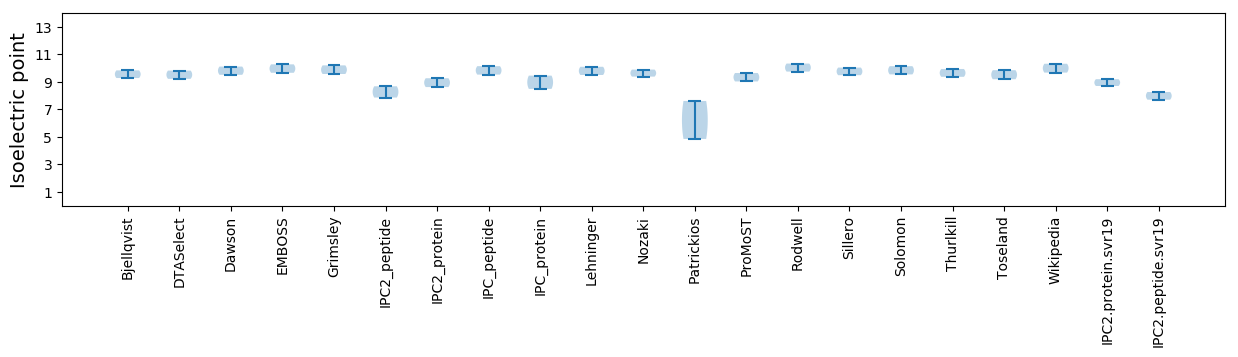
Average proteome isoelectric point is 8.95
Get precalculated fractions of proteins
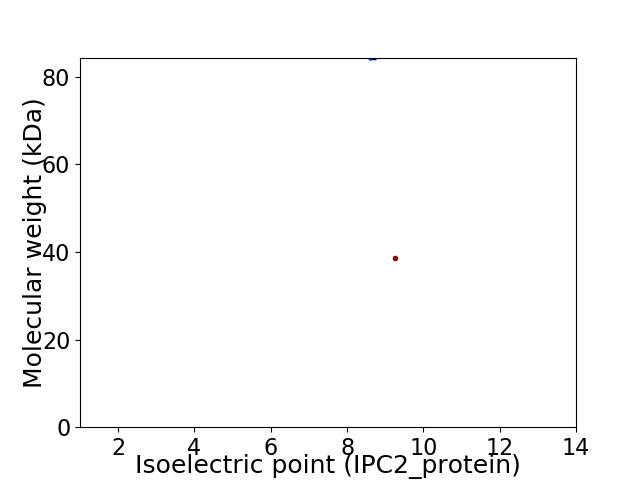
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIB8|A0A1L3KIB8_9VIRU Uncharacterized protein OS=Beihai narna-like virus 10 OX=1922437 PE=4 SV=1
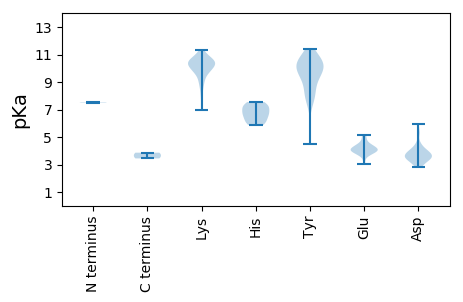
MM1 pKa = 7.51NFRR4 pKa = 11.84LEE6 pKa = 4.41DD7 pKa = 3.47QLVMYY12 pKa = 7.76RR13 pKa = 11.84TAAQWPPEE21 pKa = 3.97KK22 pKa = 10.08FIKK25 pKa = 8.98FAKK28 pKa = 8.76YY29 pKa = 4.53TTAWPMAKK37 pKa = 9.78FLEE40 pKa = 4.55NDD42 pKa = 3.58LPAVPDD48 pKa = 4.22GFQGNPLWSGAIKK61 pKa = 10.55RR62 pKa = 11.84FLKK65 pKa = 9.72TRR67 pKa = 11.84IVARR71 pKa = 11.84SPRR74 pKa = 11.84NGRR77 pKa = 11.84LFFGILQGVKK87 pKa = 9.68KK88 pKa = 10.57ACAQVPEE95 pKa = 4.4SFVMEE100 pKa = 4.69AYY102 pKa = 10.1VKK104 pKa = 10.7HH105 pKa = 6.78RR106 pKa = 11.84DD107 pKa = 3.65ALSGEE112 pKa = 4.15PRR114 pKa = 11.84GVDD117 pKa = 3.16PGQMSFYY124 pKa = 10.1YY125 pKa = 10.36RR126 pKa = 11.84EE127 pKa = 3.74FFKK130 pKa = 10.63RR131 pKa = 11.84FRR133 pKa = 11.84PRR135 pKa = 11.84KK136 pKa = 7.45PKK138 pKa = 10.4LYY140 pKa = 9.31EE141 pKa = 4.05ASNSASFEE149 pKa = 4.46TVRR152 pKa = 11.84SMGGARR158 pKa = 11.84GWIQSNHH165 pKa = 6.58DD166 pKa = 4.01LVDD169 pKa = 4.57DD170 pKa = 5.36DD171 pKa = 5.28GLIGMFEE178 pKa = 4.0TRR180 pKa = 11.84PGKK183 pKa = 10.48VEE185 pKa = 4.22TVKK188 pKa = 11.04GPALEE193 pKa = 4.15PTFDD197 pKa = 3.88EE198 pKa = 5.09LVDD201 pKa = 4.21LALEE205 pKa = 4.27EE206 pKa = 4.45PKK208 pKa = 10.37SVRR211 pKa = 11.84VGAVLEE217 pKa = 4.05PLKK220 pKa = 11.06VRR222 pKa = 11.84LITKK226 pKa = 10.13GNTLRR231 pKa = 11.84YY232 pKa = 8.16WLSRR236 pKa = 11.84DD237 pKa = 3.54CQKK240 pKa = 10.81QLWSYY245 pKa = 8.2LQKK248 pKa = 10.58YY249 pKa = 9.29DD250 pKa = 3.87QFALTGRR257 pKa = 11.84PLMAYY262 pKa = 9.04DD263 pKa = 3.39LHH265 pKa = 7.43GILIRR270 pKa = 11.84EE271 pKa = 4.38EE272 pKa = 3.95KK273 pKa = 10.76LGLKK277 pKa = 9.34FDD279 pKa = 3.65KK280 pKa = 10.14WVSGDD285 pKa = 3.39YY286 pKa = 10.71AAATDD291 pKa = 4.01TLDD294 pKa = 4.49LRR296 pKa = 11.84HH297 pKa = 6.01TKK299 pKa = 10.29AAFEE303 pKa = 4.06EE304 pKa = 4.15ALRR307 pKa = 11.84MCMFSYY313 pKa = 10.26SPKK316 pKa = 8.82YY317 pKa = 9.8QDD319 pKa = 3.12VLRR322 pKa = 11.84SVLYY326 pKa = 9.47EE327 pKa = 3.95QEE329 pKa = 3.21IHH331 pKa = 6.04YY332 pKa = 10.6PEE334 pKa = 4.86GAVKK338 pKa = 10.2KK339 pKa = 10.34CAGKK343 pKa = 10.22LDD345 pKa = 3.92PVMQRR350 pKa = 11.84TGQLMGSTLSFPILCAINLCAYY372 pKa = 7.73WAALEE377 pKa = 4.12EE378 pKa = 4.31LTGRR382 pKa = 11.84EE383 pKa = 4.01FDD385 pKa = 3.75VHH387 pKa = 6.84DD388 pKa = 5.31LPVLVNGDD396 pKa = 4.49DD397 pKa = 3.29ILFRR401 pKa = 11.84TDD403 pKa = 3.58DD404 pKa = 3.56RR405 pKa = 11.84LYY407 pKa = 10.75DD408 pKa = 2.82IWLRR412 pKa = 11.84KK413 pKa = 7.0TKK415 pKa = 10.3EE416 pKa = 3.73VGFEE420 pKa = 3.76LSLGKK425 pKa = 10.42NYY427 pKa = 9.5VHH429 pKa = 7.25PNYY432 pKa = 9.83LTVNSQLYY440 pKa = 8.53YY441 pKa = 10.86FKK443 pKa = 10.5NTTFRR448 pKa = 11.84GKK450 pKa = 10.1KK451 pKa = 9.12KK452 pKa = 10.82SLFIPLGYY460 pKa = 10.26LRR462 pKa = 11.84AGLLTGQSKK471 pKa = 8.2ITGRR475 pKa = 11.84QEE477 pKa = 3.79AKK479 pKa = 10.32AAPLWDD485 pKa = 3.69YY486 pKa = 10.76FNKK489 pKa = 8.09VTRR492 pKa = 11.84EE493 pKa = 3.78ALDD496 pKa = 4.11PIRR499 pKa = 11.84AKK501 pKa = 10.54RR502 pKa = 11.84RR503 pKa = 11.84FISYY507 pKa = 10.31HH508 pKa = 6.44KK509 pKa = 9.77DD510 pKa = 2.91TIVRR514 pKa = 11.84LTQKK518 pKa = 10.68GKK520 pKa = 9.21WNLFASPMKK529 pKa = 10.46GGLGFEE535 pKa = 4.19PVEE538 pKa = 4.09GDD540 pKa = 3.42KK541 pKa = 11.36LRR543 pKa = 11.84FTPFQRR549 pKa = 11.84RR550 pKa = 11.84WADD553 pKa = 3.37FMDD556 pKa = 3.27WKK558 pKa = 10.79LRR560 pKa = 11.84NDD562 pKa = 3.8PDD564 pKa = 3.65NFGTISLIQSRR575 pKa = 11.84SSKK578 pKa = 10.48DD579 pKa = 3.42GPRR582 pKa = 11.84VLHH585 pKa = 6.22NPQLTVQPRR594 pKa = 11.84YY595 pKa = 9.81GPYY598 pKa = 9.7EE599 pKa = 4.02EE600 pKa = 4.84GVVEE604 pKa = 5.15IKK606 pKa = 10.01DD607 pKa = 3.52TTIPLQILAARR618 pKa = 11.84LEE620 pKa = 4.37INDD623 pKa = 3.93AEE625 pKa = 4.37DD626 pKa = 3.22MKK628 pKa = 11.26SALRR632 pKa = 11.84VKK634 pKa = 10.28FPKK637 pKa = 10.33KK638 pKa = 8.64EE639 pKa = 3.98TLKK642 pKa = 10.62EE643 pKa = 3.78FRR645 pKa = 11.84SRR647 pKa = 11.84NWRR650 pKa = 11.84QLKK653 pKa = 10.61GSLTCDD659 pKa = 2.88RR660 pKa = 11.84FRR662 pKa = 11.84LMEE665 pKa = 4.67KK666 pKa = 9.81RR667 pKa = 11.84SEE669 pKa = 4.17MNSKK673 pKa = 9.84VLSALEE679 pKa = 3.87QMDD682 pKa = 4.04GVTEE686 pKa = 4.12LLQEE690 pKa = 4.27YY691 pKa = 10.35KK692 pKa = 9.74STGTFNSLSLEE703 pKa = 3.84IMNDD707 pKa = 2.88IVEE710 pKa = 5.11GICPSCSAAEE720 pKa = 3.98KK721 pKa = 10.4KK722 pKa = 10.38KK723 pKa = 9.81YY724 pKa = 9.91LSHH727 pKa = 7.23RR728 pKa = 11.84EE729 pKa = 3.79YY730 pKa = 11.38SEE732 pKa = 3.86
MM1 pKa = 7.51NFRR4 pKa = 11.84LEE6 pKa = 4.41DD7 pKa = 3.47QLVMYY12 pKa = 7.76RR13 pKa = 11.84TAAQWPPEE21 pKa = 3.97KK22 pKa = 10.08FIKK25 pKa = 8.98FAKK28 pKa = 8.76YY29 pKa = 4.53TTAWPMAKK37 pKa = 9.78FLEE40 pKa = 4.55NDD42 pKa = 3.58LPAVPDD48 pKa = 4.22GFQGNPLWSGAIKK61 pKa = 10.55RR62 pKa = 11.84FLKK65 pKa = 9.72TRR67 pKa = 11.84IVARR71 pKa = 11.84SPRR74 pKa = 11.84NGRR77 pKa = 11.84LFFGILQGVKK87 pKa = 9.68KK88 pKa = 10.57ACAQVPEE95 pKa = 4.4SFVMEE100 pKa = 4.69AYY102 pKa = 10.1VKK104 pKa = 10.7HH105 pKa = 6.78RR106 pKa = 11.84DD107 pKa = 3.65ALSGEE112 pKa = 4.15PRR114 pKa = 11.84GVDD117 pKa = 3.16PGQMSFYY124 pKa = 10.1YY125 pKa = 10.36RR126 pKa = 11.84EE127 pKa = 3.74FFKK130 pKa = 10.63RR131 pKa = 11.84FRR133 pKa = 11.84PRR135 pKa = 11.84KK136 pKa = 7.45PKK138 pKa = 10.4LYY140 pKa = 9.31EE141 pKa = 4.05ASNSASFEE149 pKa = 4.46TVRR152 pKa = 11.84SMGGARR158 pKa = 11.84GWIQSNHH165 pKa = 6.58DD166 pKa = 4.01LVDD169 pKa = 4.57DD170 pKa = 5.36DD171 pKa = 5.28GLIGMFEE178 pKa = 4.0TRR180 pKa = 11.84PGKK183 pKa = 10.48VEE185 pKa = 4.22TVKK188 pKa = 11.04GPALEE193 pKa = 4.15PTFDD197 pKa = 3.88EE198 pKa = 5.09LVDD201 pKa = 4.21LALEE205 pKa = 4.27EE206 pKa = 4.45PKK208 pKa = 10.37SVRR211 pKa = 11.84VGAVLEE217 pKa = 4.05PLKK220 pKa = 11.06VRR222 pKa = 11.84LITKK226 pKa = 10.13GNTLRR231 pKa = 11.84YY232 pKa = 8.16WLSRR236 pKa = 11.84DD237 pKa = 3.54CQKK240 pKa = 10.81QLWSYY245 pKa = 8.2LQKK248 pKa = 10.58YY249 pKa = 9.29DD250 pKa = 3.87QFALTGRR257 pKa = 11.84PLMAYY262 pKa = 9.04DD263 pKa = 3.39LHH265 pKa = 7.43GILIRR270 pKa = 11.84EE271 pKa = 4.38EE272 pKa = 3.95KK273 pKa = 10.76LGLKK277 pKa = 9.34FDD279 pKa = 3.65KK280 pKa = 10.14WVSGDD285 pKa = 3.39YY286 pKa = 10.71AAATDD291 pKa = 4.01TLDD294 pKa = 4.49LRR296 pKa = 11.84HH297 pKa = 6.01TKK299 pKa = 10.29AAFEE303 pKa = 4.06EE304 pKa = 4.15ALRR307 pKa = 11.84MCMFSYY313 pKa = 10.26SPKK316 pKa = 8.82YY317 pKa = 9.8QDD319 pKa = 3.12VLRR322 pKa = 11.84SVLYY326 pKa = 9.47EE327 pKa = 3.95QEE329 pKa = 3.21IHH331 pKa = 6.04YY332 pKa = 10.6PEE334 pKa = 4.86GAVKK338 pKa = 10.2KK339 pKa = 10.34CAGKK343 pKa = 10.22LDD345 pKa = 3.92PVMQRR350 pKa = 11.84TGQLMGSTLSFPILCAINLCAYY372 pKa = 7.73WAALEE377 pKa = 4.12EE378 pKa = 4.31LTGRR382 pKa = 11.84EE383 pKa = 4.01FDD385 pKa = 3.75VHH387 pKa = 6.84DD388 pKa = 5.31LPVLVNGDD396 pKa = 4.49DD397 pKa = 3.29ILFRR401 pKa = 11.84TDD403 pKa = 3.58DD404 pKa = 3.56RR405 pKa = 11.84LYY407 pKa = 10.75DD408 pKa = 2.82IWLRR412 pKa = 11.84KK413 pKa = 7.0TKK415 pKa = 10.3EE416 pKa = 3.73VGFEE420 pKa = 3.76LSLGKK425 pKa = 10.42NYY427 pKa = 9.5VHH429 pKa = 7.25PNYY432 pKa = 9.83LTVNSQLYY440 pKa = 8.53YY441 pKa = 10.86FKK443 pKa = 10.5NTTFRR448 pKa = 11.84GKK450 pKa = 10.1KK451 pKa = 9.12KK452 pKa = 10.82SLFIPLGYY460 pKa = 10.26LRR462 pKa = 11.84AGLLTGQSKK471 pKa = 8.2ITGRR475 pKa = 11.84QEE477 pKa = 3.79AKK479 pKa = 10.32AAPLWDD485 pKa = 3.69YY486 pKa = 10.76FNKK489 pKa = 8.09VTRR492 pKa = 11.84EE493 pKa = 3.78ALDD496 pKa = 4.11PIRR499 pKa = 11.84AKK501 pKa = 10.54RR502 pKa = 11.84RR503 pKa = 11.84FISYY507 pKa = 10.31HH508 pKa = 6.44KK509 pKa = 9.77DD510 pKa = 2.91TIVRR514 pKa = 11.84LTQKK518 pKa = 10.68GKK520 pKa = 9.21WNLFASPMKK529 pKa = 10.46GGLGFEE535 pKa = 4.19PVEE538 pKa = 4.09GDD540 pKa = 3.42KK541 pKa = 11.36LRR543 pKa = 11.84FTPFQRR549 pKa = 11.84RR550 pKa = 11.84WADD553 pKa = 3.37FMDD556 pKa = 3.27WKK558 pKa = 10.79LRR560 pKa = 11.84NDD562 pKa = 3.8PDD564 pKa = 3.65NFGTISLIQSRR575 pKa = 11.84SSKK578 pKa = 10.48DD579 pKa = 3.42GPRR582 pKa = 11.84VLHH585 pKa = 6.22NPQLTVQPRR594 pKa = 11.84YY595 pKa = 9.81GPYY598 pKa = 9.7EE599 pKa = 4.02EE600 pKa = 4.84GVVEE604 pKa = 5.15IKK606 pKa = 10.01DD607 pKa = 3.52TTIPLQILAARR618 pKa = 11.84LEE620 pKa = 4.37INDD623 pKa = 3.93AEE625 pKa = 4.37DD626 pKa = 3.22MKK628 pKa = 11.26SALRR632 pKa = 11.84VKK634 pKa = 10.28FPKK637 pKa = 10.33KK638 pKa = 8.64EE639 pKa = 3.98TLKK642 pKa = 10.62EE643 pKa = 3.78FRR645 pKa = 11.84SRR647 pKa = 11.84NWRR650 pKa = 11.84QLKK653 pKa = 10.61GSLTCDD659 pKa = 2.88RR660 pKa = 11.84FRR662 pKa = 11.84LMEE665 pKa = 4.67KK666 pKa = 9.81RR667 pKa = 11.84SEE669 pKa = 4.17MNSKK673 pKa = 9.84VLSALEE679 pKa = 3.87QMDD682 pKa = 4.04GVTEE686 pKa = 4.12LLQEE690 pKa = 4.27YY691 pKa = 10.35KK692 pKa = 9.74STGTFNSLSLEE703 pKa = 3.84IMNDD707 pKa = 2.88IVEE710 pKa = 5.11GICPSCSAAEE720 pKa = 3.98KK721 pKa = 10.4KK722 pKa = 10.38KK723 pKa = 9.81YY724 pKa = 9.91LSHH727 pKa = 7.23RR728 pKa = 11.84EE729 pKa = 3.79YY730 pKa = 11.38SEE732 pKa = 3.86
Molecular weight: 84.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIB8|A0A1L3KIB8_9VIRU Uncharacterized protein OS=Beihai narna-like virus 10 OX=1922437 PE=4 SV=1
MM1 pKa = 7.48SNNTRR6 pKa = 11.84SPIKK10 pKa = 10.09KK11 pKa = 9.77KK12 pKa = 10.55NGGSRR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84PIRR23 pKa = 11.84GARR26 pKa = 11.84LAPFRR31 pKa = 11.84GLAQSMALVPARR43 pKa = 11.84AFGGASSSHH52 pKa = 5.88SVLKK56 pKa = 10.99GLDD59 pKa = 3.36AFDD62 pKa = 3.92EE63 pKa = 4.4HH64 pKa = 7.54HH65 pKa = 6.61VALPRR70 pKa = 11.84AVANYY75 pKa = 8.44TVIRR79 pKa = 11.84TTQVLNQADD88 pKa = 3.65ATLSVFGPLAITDD101 pKa = 4.3FQAPGYY107 pKa = 7.38GTQWSNGFCVQSVDD121 pKa = 3.56PQQSINVANNAVLKK135 pKa = 9.23TFSFMGGNNTSWNGVRR151 pKa = 11.84MTPAAFTIKK160 pKa = 10.36VMNPEE165 pKa = 4.03ALQTTRR171 pKa = 11.84GAVYY175 pKa = 10.25IGRR178 pKa = 11.84AKK180 pKa = 10.18QQLNLGGNVRR190 pKa = 11.84TWNDD194 pKa = 3.0LANALVSYY202 pKa = 9.01SAPEE206 pKa = 3.5IVAAGRR212 pKa = 11.84LALRR216 pKa = 11.84GVKK219 pKa = 9.71VDD221 pKa = 3.63ALPYY225 pKa = 10.88DD226 pKa = 3.87MQSLSDD232 pKa = 3.79FRR234 pKa = 11.84SGKK237 pKa = 9.94LLQDD241 pKa = 4.13NVFTWGDD248 pKa = 3.21QSTDD252 pKa = 3.51FDD254 pKa = 3.81GFAPIFVYY262 pKa = 10.84NPDD265 pKa = 3.45QVKK268 pKa = 10.01LQILVCCEE276 pKa = 3.01WRR278 pKa = 11.84VRR280 pKa = 11.84FDD282 pKa = 3.89PEE284 pKa = 3.57NPAYY288 pKa = 10.2ASHH291 pKa = 6.96TYY293 pKa = 9.88HH294 pKa = 7.26RR295 pKa = 11.84PTSLAYY301 pKa = 7.96WDD303 pKa = 3.53RR304 pKa = 11.84VQRR307 pKa = 11.84IGNSVGNGVFDD318 pKa = 5.97LIEE321 pKa = 4.1KK322 pKa = 9.57AAPAVIGNMAQRR334 pKa = 11.84AVGRR338 pKa = 11.84IAQGVASSSVPLLMNAAVV356 pKa = 3.44
MM1 pKa = 7.48SNNTRR6 pKa = 11.84SPIKK10 pKa = 10.09KK11 pKa = 9.77KK12 pKa = 10.55NGGSRR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84PIRR23 pKa = 11.84GARR26 pKa = 11.84LAPFRR31 pKa = 11.84GLAQSMALVPARR43 pKa = 11.84AFGGASSSHH52 pKa = 5.88SVLKK56 pKa = 10.99GLDD59 pKa = 3.36AFDD62 pKa = 3.92EE63 pKa = 4.4HH64 pKa = 7.54HH65 pKa = 6.61VALPRR70 pKa = 11.84AVANYY75 pKa = 8.44TVIRR79 pKa = 11.84TTQVLNQADD88 pKa = 3.65ATLSVFGPLAITDD101 pKa = 4.3FQAPGYY107 pKa = 7.38GTQWSNGFCVQSVDD121 pKa = 3.56PQQSINVANNAVLKK135 pKa = 9.23TFSFMGGNNTSWNGVRR151 pKa = 11.84MTPAAFTIKK160 pKa = 10.36VMNPEE165 pKa = 4.03ALQTTRR171 pKa = 11.84GAVYY175 pKa = 10.25IGRR178 pKa = 11.84AKK180 pKa = 10.18QQLNLGGNVRR190 pKa = 11.84TWNDD194 pKa = 3.0LANALVSYY202 pKa = 9.01SAPEE206 pKa = 3.5IVAAGRR212 pKa = 11.84LALRR216 pKa = 11.84GVKK219 pKa = 9.71VDD221 pKa = 3.63ALPYY225 pKa = 10.88DD226 pKa = 3.87MQSLSDD232 pKa = 3.79FRR234 pKa = 11.84SGKK237 pKa = 9.94LLQDD241 pKa = 4.13NVFTWGDD248 pKa = 3.21QSTDD252 pKa = 3.51FDD254 pKa = 3.81GFAPIFVYY262 pKa = 10.84NPDD265 pKa = 3.45QVKK268 pKa = 10.01LQILVCCEE276 pKa = 3.01WRR278 pKa = 11.84VRR280 pKa = 11.84FDD282 pKa = 3.89PEE284 pKa = 3.57NPAYY288 pKa = 10.2ASHH291 pKa = 6.96TYY293 pKa = 9.88HH294 pKa = 7.26RR295 pKa = 11.84PTSLAYY301 pKa = 7.96WDD303 pKa = 3.53RR304 pKa = 11.84VQRR307 pKa = 11.84IGNSVGNGVFDD318 pKa = 5.97LIEE321 pKa = 4.1KK322 pKa = 9.57AAPAVIGNMAQRR334 pKa = 11.84AVGRR338 pKa = 11.84IAQGVASSSVPLLMNAAVV356 pKa = 3.44
Molecular weight: 38.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1088 |
356 |
732 |
544.0 |
61.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.18 ± 1.969 | 1.103 ± 0.142 |
5.423 ± 0.352 | 5.055 ± 1.834 |
5.055 ± 0.305 | 7.169 ± 0.685 |
1.379 ± 0.014 | 3.86 ± 0.039 |
6.342 ± 1.77 | 9.835 ± 1.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.482 ± 0.128 | 4.32 ± 1.318 |
5.239 ± 0.053 | 4.228 ± 0.604 |
7.169 ± 0.233 | 6.25 ± 0.573 |
5.055 ± 0.001 | 6.526 ± 1.341 |
1.838 ± 0.083 | 3.493 ± 0.525 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
