
bacterium HR36
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
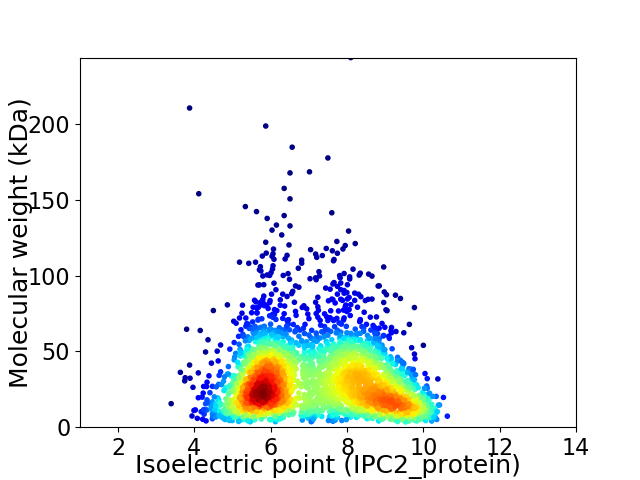
Virtual 2D-PAGE plot for 2862 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H6AEK2|A0A2H6AEK2_9BACT Uncharacterized protein OS=bacterium HR36 OX=2035431 GN=HRbin36_01500 PE=4 SV=1
MM1 pKa = 7.75PSLTVSLPSLTEE13 pKa = 3.76LHH15 pKa = 6.38EE16 pKa = 4.46GEE18 pKa = 4.48YY19 pKa = 10.92VYY21 pKa = 10.68LYY23 pKa = 10.04PDD25 pKa = 3.56VNHH28 pKa = 6.98TPQCSSQPLTFSASGLPDD46 pKa = 4.0GLSLDD51 pKa = 4.04HH52 pKa = 6.44STGLITGVLPHH63 pKa = 6.65SLANCTNPIRR73 pKa = 11.84EE74 pKa = 4.07YY75 pKa = 10.58TITLTATDD83 pKa = 5.42GIDD86 pKa = 3.36TASASATLRR95 pKa = 11.84IHH97 pKa = 5.49NTDD100 pKa = 3.26YY101 pKa = 11.22TLLSPGDD108 pKa = 3.59QSNTEE113 pKa = 3.68GDD115 pKa = 4.12YY116 pKa = 11.35VYY118 pKa = 11.4LDD120 pKa = 4.14LQSLWGSAYY129 pKa = 10.46AGLNMQAPLIFSTPSTSAPNSLPPGLSLDD158 pKa = 3.92PNTGIVSGTISSDD171 pKa = 2.63AATPNNSPYY180 pKa = 10.6SYY182 pKa = 10.73YY183 pKa = 9.8VTITLQNLADD193 pKa = 4.78GDD195 pKa = 3.96TSTISFNWAVIDD207 pKa = 4.02NPNLAEE213 pKa = 4.51PPPDD217 pKa = 4.75PGNDD221 pKa = 3.49PPPEE225 pKa = 4.2NEE227 pKa = 4.11PPTIEE232 pKa = 4.37ALSLVNVSDD241 pKa = 4.11ASRR244 pKa = 11.84NLAPRR249 pKa = 11.84TQDD252 pKa = 3.46LLQATVIGVSDD263 pKa = 4.29PDD265 pKa = 3.8GDD267 pKa = 4.0PVSVRR272 pKa = 11.84LEE274 pKa = 3.7WRR276 pKa = 11.84VNGVTVQTVEE286 pKa = 4.27GTSATFDD293 pKa = 3.73LAQPGHH299 pKa = 6.77GDD301 pKa = 2.95VGDD304 pKa = 3.84QVTVAVILDD313 pKa = 4.32DD314 pKa = 4.11GSGPIEE320 pKa = 3.97AASSNVTVANTPPVVTDD337 pKa = 3.11ILVEE341 pKa = 3.78QGYY344 pKa = 7.9QAFEE348 pKa = 4.64DD349 pKa = 3.67DD350 pKa = 3.54VYY352 pKa = 11.56YY353 pKa = 11.14YY354 pKa = 11.25LVGQRR359 pKa = 11.84AAMHH363 pKa = 6.48FLVQDD368 pKa = 3.66DD369 pKa = 6.13DD370 pKa = 3.88MDD372 pKa = 3.72QGQGNDD378 pKa = 3.4QLRR381 pKa = 11.84LEE383 pKa = 4.74VGGEE387 pKa = 3.93LPPAEE392 pKa = 4.46NGWYY396 pKa = 9.84ISDD399 pKa = 3.77YY400 pKa = 11.02QNGEE404 pKa = 4.01FFLEE408 pKa = 4.0ATIADD413 pKa = 4.57HH414 pKa = 7.33GDD416 pKa = 3.19PDD418 pKa = 4.27QPTKK422 pKa = 10.91YY423 pKa = 10.24YY424 pKa = 9.87PLNICAKK431 pKa = 9.8DD432 pKa = 3.49NYY434 pKa = 10.51DD435 pKa = 4.37AGDD438 pKa = 3.77PPIILAVTHH447 pKa = 5.66PHH449 pKa = 4.24VTRR452 pKa = 11.84ILVTPPATMAGWGTALNPYY471 pKa = 10.02LMRR474 pKa = 11.84AGRR477 pKa = 11.84RR478 pKa = 11.84EE479 pKa = 4.0TFGVRR484 pKa = 11.84IEE486 pKa = 4.2GVGFGGGVPWGGWIWWRR503 pKa = 11.84LWDD506 pKa = 5.61DD507 pKa = 4.59DD508 pKa = 5.05FADD511 pKa = 4.62SLPNQNDD518 pKa = 3.66LLQGDD523 pKa = 3.9TRR525 pKa = 11.84EE526 pKa = 4.16TVTMVGGPTGAWFVNTTFEE545 pKa = 4.24LTADD549 pKa = 3.81AASDD553 pKa = 4.36DD554 pKa = 4.0IIGRR558 pKa = 11.84DD559 pKa = 3.67GNTGAGWPEE568 pKa = 3.9GAVKK572 pKa = 10.18EE573 pKa = 4.15LYY575 pKa = 10.43FYY577 pKa = 9.97VWWWYY582 pKa = 7.79TVLWGSARR590 pKa = 11.84SPIFYY595 pKa = 10.61VEE597 pKa = 4.12WVPP600 pKa = 4.82
MM1 pKa = 7.75PSLTVSLPSLTEE13 pKa = 3.76LHH15 pKa = 6.38EE16 pKa = 4.46GEE18 pKa = 4.48YY19 pKa = 10.92VYY21 pKa = 10.68LYY23 pKa = 10.04PDD25 pKa = 3.56VNHH28 pKa = 6.98TPQCSSQPLTFSASGLPDD46 pKa = 4.0GLSLDD51 pKa = 4.04HH52 pKa = 6.44STGLITGVLPHH63 pKa = 6.65SLANCTNPIRR73 pKa = 11.84EE74 pKa = 4.07YY75 pKa = 10.58TITLTATDD83 pKa = 5.42GIDD86 pKa = 3.36TASASATLRR95 pKa = 11.84IHH97 pKa = 5.49NTDD100 pKa = 3.26YY101 pKa = 11.22TLLSPGDD108 pKa = 3.59QSNTEE113 pKa = 3.68GDD115 pKa = 4.12YY116 pKa = 11.35VYY118 pKa = 11.4LDD120 pKa = 4.14LQSLWGSAYY129 pKa = 10.46AGLNMQAPLIFSTPSTSAPNSLPPGLSLDD158 pKa = 3.92PNTGIVSGTISSDD171 pKa = 2.63AATPNNSPYY180 pKa = 10.6SYY182 pKa = 10.73YY183 pKa = 9.8VTITLQNLADD193 pKa = 4.78GDD195 pKa = 3.96TSTISFNWAVIDD207 pKa = 4.02NPNLAEE213 pKa = 4.51PPPDD217 pKa = 4.75PGNDD221 pKa = 3.49PPPEE225 pKa = 4.2NEE227 pKa = 4.11PPTIEE232 pKa = 4.37ALSLVNVSDD241 pKa = 4.11ASRR244 pKa = 11.84NLAPRR249 pKa = 11.84TQDD252 pKa = 3.46LLQATVIGVSDD263 pKa = 4.29PDD265 pKa = 3.8GDD267 pKa = 4.0PVSVRR272 pKa = 11.84LEE274 pKa = 3.7WRR276 pKa = 11.84VNGVTVQTVEE286 pKa = 4.27GTSATFDD293 pKa = 3.73LAQPGHH299 pKa = 6.77GDD301 pKa = 2.95VGDD304 pKa = 3.84QVTVAVILDD313 pKa = 4.32DD314 pKa = 4.11GSGPIEE320 pKa = 3.97AASSNVTVANTPPVVTDD337 pKa = 3.11ILVEE341 pKa = 3.78QGYY344 pKa = 7.9QAFEE348 pKa = 4.64DD349 pKa = 3.67DD350 pKa = 3.54VYY352 pKa = 11.56YY353 pKa = 11.14YY354 pKa = 11.25LVGQRR359 pKa = 11.84AAMHH363 pKa = 6.48FLVQDD368 pKa = 3.66DD369 pKa = 6.13DD370 pKa = 3.88MDD372 pKa = 3.72QGQGNDD378 pKa = 3.4QLRR381 pKa = 11.84LEE383 pKa = 4.74VGGEE387 pKa = 3.93LPPAEE392 pKa = 4.46NGWYY396 pKa = 9.84ISDD399 pKa = 3.77YY400 pKa = 11.02QNGEE404 pKa = 4.01FFLEE408 pKa = 4.0ATIADD413 pKa = 4.57HH414 pKa = 7.33GDD416 pKa = 3.19PDD418 pKa = 4.27QPTKK422 pKa = 10.91YY423 pKa = 10.24YY424 pKa = 9.87PLNICAKK431 pKa = 9.8DD432 pKa = 3.49NYY434 pKa = 10.51DD435 pKa = 4.37AGDD438 pKa = 3.77PPIILAVTHH447 pKa = 5.66PHH449 pKa = 4.24VTRR452 pKa = 11.84ILVTPPATMAGWGTALNPYY471 pKa = 10.02LMRR474 pKa = 11.84AGRR477 pKa = 11.84RR478 pKa = 11.84EE479 pKa = 4.0TFGVRR484 pKa = 11.84IEE486 pKa = 4.2GVGFGGGVPWGGWIWWRR503 pKa = 11.84LWDD506 pKa = 5.61DD507 pKa = 4.59DD508 pKa = 5.05FADD511 pKa = 4.62SLPNQNDD518 pKa = 3.66LLQGDD523 pKa = 3.9TRR525 pKa = 11.84EE526 pKa = 4.16TVTMVGGPTGAWFVNTTFEE545 pKa = 4.24LTADD549 pKa = 3.81AASDD553 pKa = 4.36DD554 pKa = 4.0IIGRR558 pKa = 11.84DD559 pKa = 3.67GNTGAGWPEE568 pKa = 3.9GAVKK572 pKa = 10.18EE573 pKa = 4.15LYY575 pKa = 10.43FYY577 pKa = 9.97VWWWYY582 pKa = 7.79TVLWGSARR590 pKa = 11.84SPIFYY595 pKa = 10.61VEE597 pKa = 4.12WVPP600 pKa = 4.82
Molecular weight: 64.7 kDa
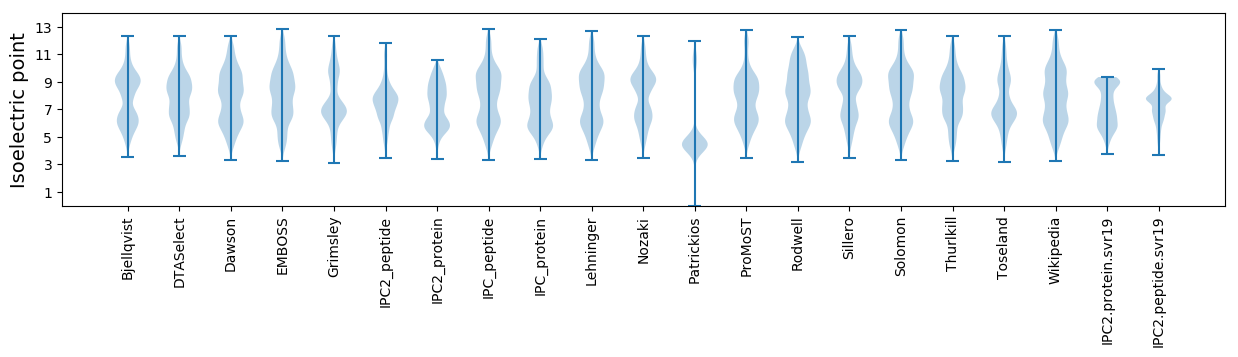
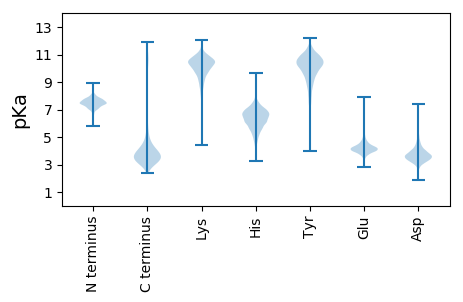
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H6ACE3|A0A2H6ACE3_9BACT Uncharacterized protein OS=bacterium HR36 OX=2035431 GN=HRbin36_00745 PE=4 SV=1
MM1 pKa = 7.42SLAFEE6 pKa = 4.29NQVVLITGAARR17 pKa = 11.84GLGRR21 pKa = 11.84QLALEE26 pKa = 4.28MALHH30 pKa = 6.27GAVIAAIDD38 pKa = 3.84RR39 pKa = 11.84DD40 pKa = 4.25VEE42 pKa = 4.19PLASLMQEE50 pKa = 3.71LKK52 pKa = 10.44EE53 pKa = 4.09RR54 pKa = 11.84GRR56 pKa = 11.84AGAFAQADD64 pKa = 3.74VTQRR68 pKa = 11.84QQLFQAVQSVEE79 pKa = 3.87QHH81 pKa = 6.33LGPVSILIANAGIGSGTSALAFDD104 pKa = 4.84PEE106 pKa = 4.23QFAEE110 pKa = 4.27MVEE113 pKa = 4.25VNLVGVANSIAAVLPGMLQRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84GHH137 pKa = 6.4LVAISSLASLRR148 pKa = 11.84GLAFLGGYY156 pKa = 8.21CASKK160 pKa = 10.85AGVNALMEE168 pKa = 4.46SLRR171 pKa = 11.84LEE173 pKa = 4.06LAPRR177 pKa = 11.84DD178 pKa = 3.64IACTIICPGWIRR190 pKa = 11.84TNLTAHH196 pKa = 7.01LPLPKK201 pKa = 9.33PDD203 pKa = 3.16IMPVEE208 pKa = 4.0EE209 pKa = 3.69AARR212 pKa = 11.84RR213 pKa = 11.84IVRR216 pKa = 11.84AIRR219 pKa = 11.84QRR221 pKa = 11.84RR222 pKa = 11.84AFYY225 pKa = 10.63AFPWSTAWILRR236 pKa = 11.84LCRR239 pKa = 11.84LLPPSLSDD247 pKa = 3.01WLLRR251 pKa = 11.84SYY253 pKa = 11.04GRR255 pKa = 11.84RR256 pKa = 11.84LQKK259 pKa = 10.44KK260 pKa = 8.51LEE262 pKa = 4.12RR263 pKa = 11.84FTRR266 pKa = 11.84RR267 pKa = 3.0
MM1 pKa = 7.42SLAFEE6 pKa = 4.29NQVVLITGAARR17 pKa = 11.84GLGRR21 pKa = 11.84QLALEE26 pKa = 4.28MALHH30 pKa = 6.27GAVIAAIDD38 pKa = 3.84RR39 pKa = 11.84DD40 pKa = 4.25VEE42 pKa = 4.19PLASLMQEE50 pKa = 3.71LKK52 pKa = 10.44EE53 pKa = 4.09RR54 pKa = 11.84GRR56 pKa = 11.84AGAFAQADD64 pKa = 3.74VTQRR68 pKa = 11.84QQLFQAVQSVEE79 pKa = 3.87QHH81 pKa = 6.33LGPVSILIANAGIGSGTSALAFDD104 pKa = 4.84PEE106 pKa = 4.23QFAEE110 pKa = 4.27MVEE113 pKa = 4.25VNLVGVANSIAAVLPGMLQRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84GHH137 pKa = 6.4LVAISSLASLRR148 pKa = 11.84GLAFLGGYY156 pKa = 8.21CASKK160 pKa = 10.85AGVNALMEE168 pKa = 4.46SLRR171 pKa = 11.84LEE173 pKa = 4.06LAPRR177 pKa = 11.84DD178 pKa = 3.64IACTIICPGWIRR190 pKa = 11.84TNLTAHH196 pKa = 7.01LPLPKK201 pKa = 9.33PDD203 pKa = 3.16IMPVEE208 pKa = 4.0EE209 pKa = 3.69AARR212 pKa = 11.84RR213 pKa = 11.84IVRR216 pKa = 11.84AIRR219 pKa = 11.84QRR221 pKa = 11.84RR222 pKa = 11.84AFYY225 pKa = 10.63AFPWSTAWILRR236 pKa = 11.84LCRR239 pKa = 11.84LLPPSLSDD247 pKa = 3.01WLLRR251 pKa = 11.84SYY253 pKa = 11.04GRR255 pKa = 11.84RR256 pKa = 11.84LQKK259 pKa = 10.44KK260 pKa = 8.51LEE262 pKa = 4.12RR263 pKa = 11.84FTRR266 pKa = 11.84RR267 pKa = 3.0
Molecular weight: 29.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
869442 |
32 |
2200 |
303.8 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.132 ± 0.047 | 1.367 ± 0.019 |
4.601 ± 0.034 | 6.235 ± 0.05 |
3.434 ± 0.025 | 7.435 ± 0.04 |
2.536 ± 0.025 | 4.491 ± 0.032 |
3.021 ± 0.036 | 11.348 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.77 ± 0.018 | 2.57 ± 0.031 |
6.219 ± 0.044 | 4.9 ± 0.037 |
8.253 ± 0.044 | 5.02 ± 0.036 |
4.761 ± 0.038 | 7.319 ± 0.03 |
1.945 ± 0.026 | 2.643 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
