
Afipia sp. GAS231
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Afipia; unclassified Afipia
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
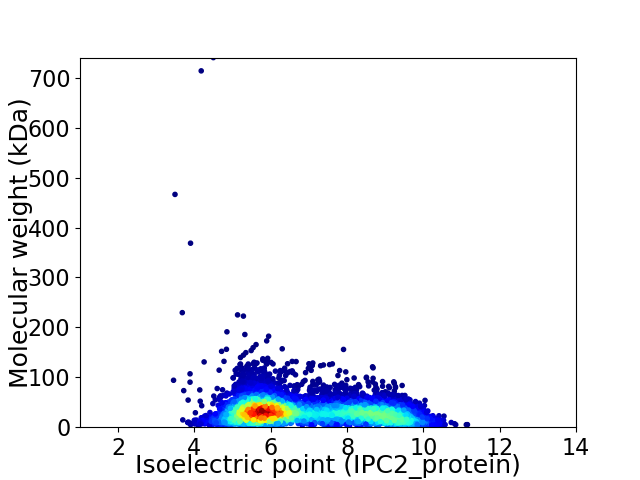
Virtual 2D-PAGE plot for 6976 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0J3I5|A0A1H0J3I5_9BRAD Tripartite-type tricarboxylate transporter receptor component TctC OS=Afipia sp. GAS231 OX=1882747 GN=SAMN05444050_3993 PE=3 SV=1
MM1 pKa = 7.66ASDD4 pKa = 4.94LPAFPGFPSPPSPPSPPPGGGTPGLIIALPFLDD37 pKa = 4.74TPPSQATDD45 pKa = 3.22SNGAANAVAEE55 pKa = 4.48GAAVNTLVGITAHH68 pKa = 6.56SVDD71 pKa = 4.76AEE73 pKa = 4.45GQTVTYY79 pKa = 10.72SLGADD84 pKa = 3.27SSGGGFKK91 pKa = 9.65IDD93 pKa = 3.64PATGVVSVADD103 pKa = 3.39SGKK106 pKa = 10.0IDD108 pKa = 4.16FEE110 pKa = 4.44TAPGHH115 pKa = 6.83AYY117 pKa = 9.24TITVNSSDD125 pKa = 3.59GFISGSQNFTIAVTDD140 pKa = 3.68VAPTAPVDD148 pKa = 4.01SNAAANAVTEE158 pKa = 4.27GAAANTTVGVTAASTDD174 pKa = 3.35INGPAVTYY182 pKa = 10.87SLTADD187 pKa = 3.34TSGGGFKK194 pKa = 9.64IDD196 pKa = 3.69PVTGIVTVADD206 pKa = 3.54ATKK209 pKa = 9.84IDD211 pKa = 4.08YY212 pKa = 10.41EE213 pKa = 4.42SAPGHH218 pKa = 6.83AYY220 pKa = 8.76TVTAQASDD228 pKa = 3.56GTLTNSQTFTIAVNDD243 pKa = 3.95VPLSTPVDD251 pKa = 3.58TNAAANNVAEE261 pKa = 4.64GAANGTAAGITAFAFDD277 pKa = 4.73PNGPATTYY285 pKa = 11.16SLTGDD290 pKa = 3.45TSGGGFTINAVTGVVTVADD309 pKa = 4.09FTKK312 pKa = 9.95IDD314 pKa = 4.01YY315 pKa = 8.72EE316 pKa = 4.57SSGAGHH322 pKa = 7.07SYY324 pKa = 10.3TITVQANDD332 pKa = 3.64GLQTTSHH339 pKa = 6.38GFTIGVADD347 pKa = 4.13VAPSTPVDD355 pKa = 3.28SDD357 pKa = 3.52AAANSVLEE365 pKa = 4.23GAANGSTVGVTASSTDD381 pKa = 3.33VNGPAVTYY389 pKa = 10.68SLTGDD394 pKa = 3.4TSGGGFTINAATGVVTVADD413 pKa = 3.81STKK416 pKa = 9.88IDD418 pKa = 3.7YY419 pKa = 10.45EE420 pKa = 4.36SAPGHH425 pKa = 6.83AYY427 pKa = 8.76TVTAQASDD435 pKa = 3.56GTLTNSQTFTIAVANVPPSTPVDD458 pKa = 3.92SNAGANTVAEE468 pKa = 4.54GAAAGSTVGVTASSTDD484 pKa = 3.18INGPAVTYY492 pKa = 10.77SLTGDD497 pKa = 3.4TSGGGFTINAVTGVVTVADD516 pKa = 3.88PTKK519 pKa = 10.02IDD521 pKa = 3.55YY522 pKa = 10.47EE523 pKa = 4.44SAPGHH528 pKa = 6.83AYY530 pKa = 8.76TVTAQASDD538 pKa = 3.69GTASSSQIFIIAVTDD553 pKa = 3.62VAPSTPVDD561 pKa = 3.28SDD563 pKa = 3.52AAANSIAEE571 pKa = 4.37GPANGSTVGVTASSTDD587 pKa = 3.33VNGPAVTYY595 pKa = 10.68SLTGDD600 pKa = 3.4TSGGGFTINATTGVITVADD619 pKa = 3.82STKK622 pKa = 9.94IDD624 pKa = 3.96YY625 pKa = 9.1EE626 pKa = 4.49SSGAGHH632 pKa = 7.06SYY634 pKa = 9.88TVTATASDD642 pKa = 4.06GTQSNSQAFVIDD654 pKa = 3.78VTDD657 pKa = 3.72VAPSTPVDD665 pKa = 3.72SNAAANQVSQGAPVGTVVGLTASSTDD691 pKa = 3.33VNGPAVTYY699 pKa = 10.63SLIGDD704 pKa = 3.99TSGGGFTINAATGVVTVADD723 pKa = 3.94PSKK726 pKa = 11.02VIFNAGSPSFDD737 pKa = 3.2VRR739 pKa = 11.84VDD741 pKa = 3.48SSDD744 pKa = 3.42GTLHH748 pKa = 5.76NQQTFTINVLQNTPPVANDD767 pKa = 3.51DD768 pKa = 3.9SLAATEE774 pKa = 5.29AGGLNNSIAGSNPAGNVILGTGSAGAVQDD803 pKa = 4.3TDD805 pKa = 3.8AQDD808 pKa = 3.51PSNALTVVAVHH819 pKa = 6.59TGPEE823 pKa = 4.3GASTGTGTVNSATVGSHH840 pKa = 5.5GTLTLNSDD848 pKa = 3.22GSYY851 pKa = 10.19TYY853 pKa = 11.06LVNQTDD859 pKa = 3.68AQVQALHH866 pKa = 6.46TSAEE870 pKa = 4.36TITDD874 pKa = 3.45VFNYY878 pKa = 8.64TIQDD882 pKa = 3.41TGGLQDD888 pKa = 3.56TATLTVTIHH897 pKa = 6.44GADD900 pKa = 4.5DD901 pKa = 4.11LPQAVADD908 pKa = 3.96TGTMTEE914 pKa = 4.62DD915 pKa = 4.24DD916 pKa = 4.68APTSFNVIANDD927 pKa = 3.86TQDD930 pKa = 3.88PDD932 pKa = 3.49HH933 pKa = 6.6TASNTITLGPGGVTVTGPGGEE954 pKa = 4.34TFANTDD960 pKa = 2.98ATASIVANQIQITLNNAHH978 pKa = 6.42FQQLALGEE986 pKa = 4.55HH987 pKa = 5.99ATITAPYY994 pKa = 8.91TLTGDD999 pKa = 3.54AGEE1002 pKa = 4.22TSSANLVVTVNGVNDD1017 pKa = 3.47VPVAVDD1023 pKa = 3.32DD1024 pKa = 4.2TGTMTEE1030 pKa = 4.5DD1031 pKa = 3.39QSGHH1035 pKa = 6.32AFTVLANDD1043 pKa = 4.33TLDD1046 pKa = 4.38ADD1048 pKa = 4.0HH1049 pKa = 7.03GAPNNVTVNAATAVTNLSAPSGEE1072 pKa = 4.92GITASDD1078 pKa = 3.05ISVSVNGSNQVVVNLGANFQHH1099 pKa = 6.09MQDD1102 pKa = 4.04GQSSTFDD1109 pKa = 3.18VAYY1112 pKa = 8.94TLHH1115 pKa = 7.18GDD1117 pKa = 3.7QPGDD1121 pKa = 3.12TSTAEE1126 pKa = 3.77LHH1128 pKa = 5.29VTVTGANDD1136 pKa = 3.3APVAGNFTFNGVNSAIGNTDD1156 pKa = 3.56LVVNDD1161 pKa = 4.82PADD1164 pKa = 3.71TAQTATGPAKK1174 pKa = 9.98TISASLLSGASDD1186 pKa = 3.43VDD1188 pKa = 3.96GPGPLIVVAGTIATAHH1204 pKa = 6.32GGAVTMQSDD1213 pKa = 3.71GDD1215 pKa = 4.34FIYY1218 pKa = 10.36KK1219 pKa = 10.23PPVGYY1224 pKa = 8.84TGSDD1228 pKa = 3.13SFNYY1232 pKa = 9.35QVSDD1236 pKa = 3.68QNAGASGVGLGTGTVTINVASPHH1259 pKa = 4.38VWYY1262 pKa = 11.05VNADD1266 pKa = 3.29AATDD1270 pKa = 3.88GDD1272 pKa = 4.25GSSEE1276 pKa = 4.07NPFNSLSHH1284 pKa = 6.38FNGAGGVDD1292 pKa = 3.72GSGDD1296 pKa = 3.88TIVLEE1301 pKa = 4.46TASAHH1306 pKa = 4.28YY1307 pKa = 9.57TGGLTLEE1314 pKa = 4.48NNEE1317 pKa = 3.97QLISQSAGVTINGTTLLAASGANAVVDD1344 pKa = 3.79GGVILGSGNNIQGVDD1359 pKa = 4.38FGATSGFAVSGSSVGTVHH1377 pKa = 7.74IDD1379 pKa = 3.35DD1380 pKa = 4.76SKK1382 pKa = 11.82SGVINNASGGGISIGGSSNVLNVDD1406 pKa = 4.21LSSVTTGGGTNGIVINGASGSFHH1429 pKa = 6.71AHH1431 pKa = 6.58GGTISGASGSDD1442 pKa = 3.35VSLSSGTLNFTDD1454 pKa = 5.21DD1455 pKa = 4.49GSISDD1460 pKa = 3.73NSGSNVSVSSMTGGTQSFTGAISGNGSTGISLTSNTGSTIDD1501 pKa = 3.57FSGGINLSTGASNAFTATGGGTVNVTGTNHH1531 pKa = 6.39LTTTTGTALNINNTTIGSSNVTFHH1555 pKa = 7.67DD1556 pKa = 3.59ISANGAANGIVLNNTGTSGHH1576 pKa = 6.56LAVTGGGSSALGGDD1590 pKa = 3.52NSGGTIQNTTGDD1602 pKa = 4.55GISLTNTSGVTFTNIHH1618 pKa = 4.78VTNTSGDD1625 pKa = 4.13GINGTTVGGFTLANSTVDD1643 pKa = 3.46GANGGSAVNQGSIYY1657 pKa = 9.62FTGLTGTANVTNDD1670 pKa = 3.49VIKK1673 pKa = 10.71GGHH1676 pKa = 5.55QDD1678 pKa = 3.21NVSILTSSGTLDD1690 pKa = 3.47LTVSGSTIRR1699 pKa = 11.84DD1700 pKa = 3.21TDD1702 pKa = 3.14TGTNGNDD1709 pKa = 3.23NLFIEE1714 pKa = 5.06ANGTSTVTAHH1724 pKa = 5.45VQNNTFAATNGDD1736 pKa = 3.63HH1737 pKa = 6.43LQTVGDD1743 pKa = 4.88DD1744 pKa = 3.55SATLNIVATGNTLSGGGGVNALGEE1768 pKa = 4.86GITISGGHH1776 pKa = 6.06SSPSPSTEE1784 pKa = 3.2HH1785 pKa = 6.47VNFNISNNTMTGTIQGGAINVNEE1808 pKa = 4.38GAGSGTWQGQINNNTVGNPAITDD1831 pKa = 3.42SGAAQSSAIRR1841 pKa = 11.84AEE1843 pKa = 3.91NHH1845 pKa = 6.27SNGGALTVLIDD1856 pKa = 4.3GNNLSQFNGVGISLQDD1872 pKa = 3.87GEE1874 pKa = 5.09SSTSTATLNATVTNNVEE1891 pKa = 4.46SNPGSTFNGGPNYY1904 pKa = 10.7GLLLNVGVASGSSNVAHH1921 pKa = 7.16ANIHH1925 pKa = 6.86DD1926 pKa = 4.01NTLVASTSGGFGIRR1940 pKa = 11.84VRR1942 pKa = 11.84EE1943 pKa = 3.95RR1944 pKa = 11.84FGADD1948 pKa = 2.47IFLDD1952 pKa = 3.88NYY1954 pKa = 11.14AGGSTDD1960 pKa = 3.21QTAVANYY1967 pKa = 9.52LVAKK1971 pKa = 10.39NPGNTAVAAMSLGGGATQGGGFFNGTTPTPIVPLQLFAAGGVQASPTTPGEE2022 pKa = 4.15MNLTMSEE2029 pKa = 4.25LNVIVAAAIADD2040 pKa = 3.65WTAAGASAAQIDD2052 pKa = 4.23AMHH2055 pKa = 6.95AASFSIANLSDD2066 pKa = 3.26GVLGQEE2072 pKa = 4.16TPGAIQVSSNAAGNGWFVDD2091 pKa = 3.6ATPAGNSEE2099 pKa = 4.48FPHH2102 pKa = 6.52AANPADD2108 pKa = 4.19TDD2110 pKa = 4.49LLTDD2114 pKa = 4.57PSNAAAGHH2122 pKa = 5.78VDD2124 pKa = 3.8LLTVVMHH2131 pKa = 6.43EE2132 pKa = 4.46LGHH2135 pKa = 7.0IIGIDD2140 pKa = 3.71DD2141 pKa = 3.57TTAPADD2147 pKa = 3.6INDD2150 pKa = 3.83LMFMYY2155 pKa = 10.59LADD2158 pKa = 3.9GEE2160 pKa = 4.62RR2161 pKa = 11.84RR2162 pKa = 11.84LPTASDD2168 pKa = 3.34VGKK2171 pKa = 9.99ADD2173 pKa = 3.7PMSSFQAVDD2182 pKa = 3.65PLAPPPPAPASTTTVAAPAGSQTFDD2207 pKa = 3.61AGHH2210 pKa = 6.83GGQTLVATAGPDD2222 pKa = 3.16TFVFANVDD2230 pKa = 3.24VHH2232 pKa = 7.49APAPPPVTLIEE2243 pKa = 4.47NYY2245 pKa = 10.67SFAQGDD2251 pKa = 3.75KK2252 pKa = 10.6FDD2254 pKa = 4.82FSAITSQFHH2263 pKa = 6.67ASGFGDD2269 pKa = 3.69ALIVRR2274 pKa = 11.84AVEE2277 pKa = 3.86DD2278 pKa = 3.68AGGKK2282 pKa = 9.19FATLQVDD2289 pKa = 3.49TLDD2292 pKa = 3.66PHH2294 pKa = 7.07GLPGMANWVNVAQLDD2309 pKa = 4.13GAHH2312 pKa = 6.88AGDD2315 pKa = 4.22AVNVLIDD2322 pKa = 3.67GQAVHH2327 pKa = 7.15LAQIHH2332 pKa = 5.76VGLLAA2337 pKa = 4.61
MM1 pKa = 7.66ASDD4 pKa = 4.94LPAFPGFPSPPSPPSPPPGGGTPGLIIALPFLDD37 pKa = 4.74TPPSQATDD45 pKa = 3.22SNGAANAVAEE55 pKa = 4.48GAAVNTLVGITAHH68 pKa = 6.56SVDD71 pKa = 4.76AEE73 pKa = 4.45GQTVTYY79 pKa = 10.72SLGADD84 pKa = 3.27SSGGGFKK91 pKa = 9.65IDD93 pKa = 3.64PATGVVSVADD103 pKa = 3.39SGKK106 pKa = 10.0IDD108 pKa = 4.16FEE110 pKa = 4.44TAPGHH115 pKa = 6.83AYY117 pKa = 9.24TITVNSSDD125 pKa = 3.59GFISGSQNFTIAVTDD140 pKa = 3.68VAPTAPVDD148 pKa = 4.01SNAAANAVTEE158 pKa = 4.27GAAANTTVGVTAASTDD174 pKa = 3.35INGPAVTYY182 pKa = 10.87SLTADD187 pKa = 3.34TSGGGFKK194 pKa = 9.64IDD196 pKa = 3.69PVTGIVTVADD206 pKa = 3.54ATKK209 pKa = 9.84IDD211 pKa = 4.08YY212 pKa = 10.41EE213 pKa = 4.42SAPGHH218 pKa = 6.83AYY220 pKa = 8.76TVTAQASDD228 pKa = 3.56GTLTNSQTFTIAVNDD243 pKa = 3.95VPLSTPVDD251 pKa = 3.58TNAAANNVAEE261 pKa = 4.64GAANGTAAGITAFAFDD277 pKa = 4.73PNGPATTYY285 pKa = 11.16SLTGDD290 pKa = 3.45TSGGGFTINAVTGVVTVADD309 pKa = 4.09FTKK312 pKa = 9.95IDD314 pKa = 4.01YY315 pKa = 8.72EE316 pKa = 4.57SSGAGHH322 pKa = 7.07SYY324 pKa = 10.3TITVQANDD332 pKa = 3.64GLQTTSHH339 pKa = 6.38GFTIGVADD347 pKa = 4.13VAPSTPVDD355 pKa = 3.28SDD357 pKa = 3.52AAANSVLEE365 pKa = 4.23GAANGSTVGVTASSTDD381 pKa = 3.33VNGPAVTYY389 pKa = 10.68SLTGDD394 pKa = 3.4TSGGGFTINAATGVVTVADD413 pKa = 3.81STKK416 pKa = 9.88IDD418 pKa = 3.7YY419 pKa = 10.45EE420 pKa = 4.36SAPGHH425 pKa = 6.83AYY427 pKa = 8.76TVTAQASDD435 pKa = 3.56GTLTNSQTFTIAVANVPPSTPVDD458 pKa = 3.92SNAGANTVAEE468 pKa = 4.54GAAAGSTVGVTASSTDD484 pKa = 3.18INGPAVTYY492 pKa = 10.77SLTGDD497 pKa = 3.4TSGGGFTINAVTGVVTVADD516 pKa = 3.88PTKK519 pKa = 10.02IDD521 pKa = 3.55YY522 pKa = 10.47EE523 pKa = 4.44SAPGHH528 pKa = 6.83AYY530 pKa = 8.76TVTAQASDD538 pKa = 3.69GTASSSQIFIIAVTDD553 pKa = 3.62VAPSTPVDD561 pKa = 3.28SDD563 pKa = 3.52AAANSIAEE571 pKa = 4.37GPANGSTVGVTASSTDD587 pKa = 3.33VNGPAVTYY595 pKa = 10.68SLTGDD600 pKa = 3.4TSGGGFTINATTGVITVADD619 pKa = 3.82STKK622 pKa = 9.94IDD624 pKa = 3.96YY625 pKa = 9.1EE626 pKa = 4.49SSGAGHH632 pKa = 7.06SYY634 pKa = 9.88TVTATASDD642 pKa = 4.06GTQSNSQAFVIDD654 pKa = 3.78VTDD657 pKa = 3.72VAPSTPVDD665 pKa = 3.72SNAAANQVSQGAPVGTVVGLTASSTDD691 pKa = 3.33VNGPAVTYY699 pKa = 10.63SLIGDD704 pKa = 3.99TSGGGFTINAATGVVTVADD723 pKa = 3.94PSKK726 pKa = 11.02VIFNAGSPSFDD737 pKa = 3.2VRR739 pKa = 11.84VDD741 pKa = 3.48SSDD744 pKa = 3.42GTLHH748 pKa = 5.76NQQTFTINVLQNTPPVANDD767 pKa = 3.51DD768 pKa = 3.9SLAATEE774 pKa = 5.29AGGLNNSIAGSNPAGNVILGTGSAGAVQDD803 pKa = 4.3TDD805 pKa = 3.8AQDD808 pKa = 3.51PSNALTVVAVHH819 pKa = 6.59TGPEE823 pKa = 4.3GASTGTGTVNSATVGSHH840 pKa = 5.5GTLTLNSDD848 pKa = 3.22GSYY851 pKa = 10.19TYY853 pKa = 11.06LVNQTDD859 pKa = 3.68AQVQALHH866 pKa = 6.46TSAEE870 pKa = 4.36TITDD874 pKa = 3.45VFNYY878 pKa = 8.64TIQDD882 pKa = 3.41TGGLQDD888 pKa = 3.56TATLTVTIHH897 pKa = 6.44GADD900 pKa = 4.5DD901 pKa = 4.11LPQAVADD908 pKa = 3.96TGTMTEE914 pKa = 4.62DD915 pKa = 4.24DD916 pKa = 4.68APTSFNVIANDD927 pKa = 3.86TQDD930 pKa = 3.88PDD932 pKa = 3.49HH933 pKa = 6.6TASNTITLGPGGVTVTGPGGEE954 pKa = 4.34TFANTDD960 pKa = 2.98ATASIVANQIQITLNNAHH978 pKa = 6.42FQQLALGEE986 pKa = 4.55HH987 pKa = 5.99ATITAPYY994 pKa = 8.91TLTGDD999 pKa = 3.54AGEE1002 pKa = 4.22TSSANLVVTVNGVNDD1017 pKa = 3.47VPVAVDD1023 pKa = 3.32DD1024 pKa = 4.2TGTMTEE1030 pKa = 4.5DD1031 pKa = 3.39QSGHH1035 pKa = 6.32AFTVLANDD1043 pKa = 4.33TLDD1046 pKa = 4.38ADD1048 pKa = 4.0HH1049 pKa = 7.03GAPNNVTVNAATAVTNLSAPSGEE1072 pKa = 4.92GITASDD1078 pKa = 3.05ISVSVNGSNQVVVNLGANFQHH1099 pKa = 6.09MQDD1102 pKa = 4.04GQSSTFDD1109 pKa = 3.18VAYY1112 pKa = 8.94TLHH1115 pKa = 7.18GDD1117 pKa = 3.7QPGDD1121 pKa = 3.12TSTAEE1126 pKa = 3.77LHH1128 pKa = 5.29VTVTGANDD1136 pKa = 3.3APVAGNFTFNGVNSAIGNTDD1156 pKa = 3.56LVVNDD1161 pKa = 4.82PADD1164 pKa = 3.71TAQTATGPAKK1174 pKa = 9.98TISASLLSGASDD1186 pKa = 3.43VDD1188 pKa = 3.96GPGPLIVVAGTIATAHH1204 pKa = 6.32GGAVTMQSDD1213 pKa = 3.71GDD1215 pKa = 4.34FIYY1218 pKa = 10.36KK1219 pKa = 10.23PPVGYY1224 pKa = 8.84TGSDD1228 pKa = 3.13SFNYY1232 pKa = 9.35QVSDD1236 pKa = 3.68QNAGASGVGLGTGTVTINVASPHH1259 pKa = 4.38VWYY1262 pKa = 11.05VNADD1266 pKa = 3.29AATDD1270 pKa = 3.88GDD1272 pKa = 4.25GSSEE1276 pKa = 4.07NPFNSLSHH1284 pKa = 6.38FNGAGGVDD1292 pKa = 3.72GSGDD1296 pKa = 3.88TIVLEE1301 pKa = 4.46TASAHH1306 pKa = 4.28YY1307 pKa = 9.57TGGLTLEE1314 pKa = 4.48NNEE1317 pKa = 3.97QLISQSAGVTINGTTLLAASGANAVVDD1344 pKa = 3.79GGVILGSGNNIQGVDD1359 pKa = 4.38FGATSGFAVSGSSVGTVHH1377 pKa = 7.74IDD1379 pKa = 3.35DD1380 pKa = 4.76SKK1382 pKa = 11.82SGVINNASGGGISIGGSSNVLNVDD1406 pKa = 4.21LSSVTTGGGTNGIVINGASGSFHH1429 pKa = 6.71AHH1431 pKa = 6.58GGTISGASGSDD1442 pKa = 3.35VSLSSGTLNFTDD1454 pKa = 5.21DD1455 pKa = 4.49GSISDD1460 pKa = 3.73NSGSNVSVSSMTGGTQSFTGAISGNGSTGISLTSNTGSTIDD1501 pKa = 3.57FSGGINLSTGASNAFTATGGGTVNVTGTNHH1531 pKa = 6.39LTTTTGTALNINNTTIGSSNVTFHH1555 pKa = 7.67DD1556 pKa = 3.59ISANGAANGIVLNNTGTSGHH1576 pKa = 6.56LAVTGGGSSALGGDD1590 pKa = 3.52NSGGTIQNTTGDD1602 pKa = 4.55GISLTNTSGVTFTNIHH1618 pKa = 4.78VTNTSGDD1625 pKa = 4.13GINGTTVGGFTLANSTVDD1643 pKa = 3.46GANGGSAVNQGSIYY1657 pKa = 9.62FTGLTGTANVTNDD1670 pKa = 3.49VIKK1673 pKa = 10.71GGHH1676 pKa = 5.55QDD1678 pKa = 3.21NVSILTSSGTLDD1690 pKa = 3.47LTVSGSTIRR1699 pKa = 11.84DD1700 pKa = 3.21TDD1702 pKa = 3.14TGTNGNDD1709 pKa = 3.23NLFIEE1714 pKa = 5.06ANGTSTVTAHH1724 pKa = 5.45VQNNTFAATNGDD1736 pKa = 3.63HH1737 pKa = 6.43LQTVGDD1743 pKa = 4.88DD1744 pKa = 3.55SATLNIVATGNTLSGGGGVNALGEE1768 pKa = 4.86GITISGGHH1776 pKa = 6.06SSPSPSTEE1784 pKa = 3.2HH1785 pKa = 6.47VNFNISNNTMTGTIQGGAINVNEE1808 pKa = 4.38GAGSGTWQGQINNNTVGNPAITDD1831 pKa = 3.42SGAAQSSAIRR1841 pKa = 11.84AEE1843 pKa = 3.91NHH1845 pKa = 6.27SNGGALTVLIDD1856 pKa = 4.3GNNLSQFNGVGISLQDD1872 pKa = 3.87GEE1874 pKa = 5.09SSTSTATLNATVTNNVEE1891 pKa = 4.46SNPGSTFNGGPNYY1904 pKa = 10.7GLLLNVGVASGSSNVAHH1921 pKa = 7.16ANIHH1925 pKa = 6.86DD1926 pKa = 4.01NTLVASTSGGFGIRR1940 pKa = 11.84VRR1942 pKa = 11.84EE1943 pKa = 3.95RR1944 pKa = 11.84FGADD1948 pKa = 2.47IFLDD1952 pKa = 3.88NYY1954 pKa = 11.14AGGSTDD1960 pKa = 3.21QTAVANYY1967 pKa = 9.52LVAKK1971 pKa = 10.39NPGNTAVAAMSLGGGATQGGGFFNGTTPTPIVPLQLFAAGGVQASPTTPGEE2022 pKa = 4.15MNLTMSEE2029 pKa = 4.25LNVIVAAAIADD2040 pKa = 3.65WTAAGASAAQIDD2052 pKa = 4.23AMHH2055 pKa = 6.95AASFSIANLSDD2066 pKa = 3.26GVLGQEE2072 pKa = 4.16TPGAIQVSSNAAGNGWFVDD2091 pKa = 3.6ATPAGNSEE2099 pKa = 4.48FPHH2102 pKa = 6.52AANPADD2108 pKa = 4.19TDD2110 pKa = 4.49LLTDD2114 pKa = 4.57PSNAAAGHH2122 pKa = 5.78VDD2124 pKa = 3.8LLTVVMHH2131 pKa = 6.43EE2132 pKa = 4.46LGHH2135 pKa = 7.0IIGIDD2140 pKa = 3.71DD2141 pKa = 3.57TTAPADD2147 pKa = 3.6INDD2150 pKa = 3.83LMFMYY2155 pKa = 10.59LADD2158 pKa = 3.9GEE2160 pKa = 4.62RR2161 pKa = 11.84RR2162 pKa = 11.84LPTASDD2168 pKa = 3.34VGKK2171 pKa = 9.99ADD2173 pKa = 3.7PMSSFQAVDD2182 pKa = 3.65PLAPPPPAPASTTTVAAPAGSQTFDD2207 pKa = 3.61AGHH2210 pKa = 6.83GGQTLVATAGPDD2222 pKa = 3.16TFVFANVDD2230 pKa = 3.24VHH2232 pKa = 7.49APAPPPVTLIEE2243 pKa = 4.47NYY2245 pKa = 10.67SFAQGDD2251 pKa = 3.75KK2252 pKa = 10.6FDD2254 pKa = 4.82FSAITSQFHH2263 pKa = 6.67ASGFGDD2269 pKa = 3.69ALIVRR2274 pKa = 11.84AVEE2277 pKa = 3.86DD2278 pKa = 3.68AGGKK2282 pKa = 9.19FATLQVDD2289 pKa = 3.49TLDD2292 pKa = 3.66PHH2294 pKa = 7.07GLPGMANWVNVAQLDD2309 pKa = 4.13GAHH2312 pKa = 6.88AGDD2315 pKa = 4.22AVNVLIDD2322 pKa = 3.67GQAVHH2327 pKa = 7.15LAQIHH2332 pKa = 5.76VGLLAA2337 pKa = 4.61
Molecular weight: 229.47 kDa
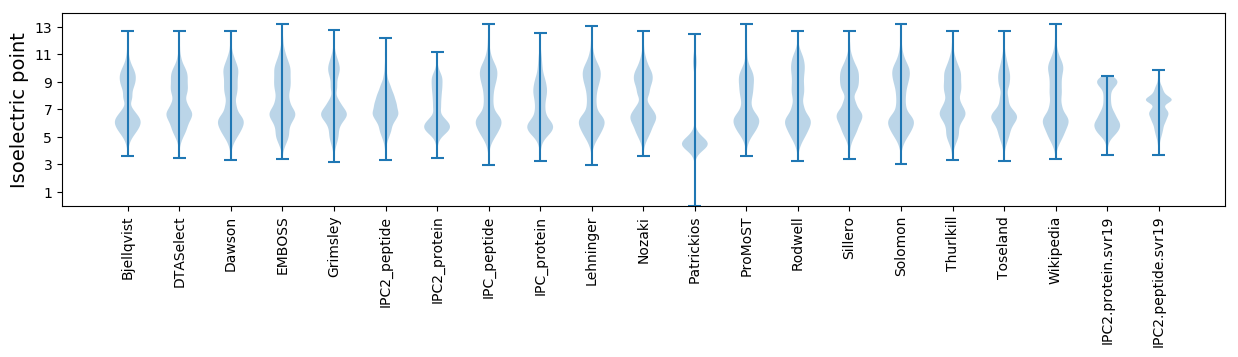
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0G2K8|A0A1H0G2K8_9BRAD Uncharacterized protein OS=Afipia sp. GAS231 OX=1882747 GN=SAMN05444050_2993 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATTGGRR28 pKa = 11.84KK29 pKa = 9.06VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATTGGRR28 pKa = 11.84KK29 pKa = 9.06VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2166649 |
29 |
7515 |
310.6 |
33.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.362 ± 0.037 | 0.863 ± 0.01 |
5.483 ± 0.025 | 5.152 ± 0.032 |
3.855 ± 0.02 | 8.507 ± 0.047 |
2.016 ± 0.014 | 5.417 ± 0.022 |
3.794 ± 0.031 | 9.678 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.481 ± 0.016 | 2.935 ± 0.035 |
5.249 ± 0.031 | 3.152 ± 0.014 |
6.647 ± 0.043 | 5.766 ± 0.032 |
5.546 ± 0.047 | 7.465 ± 0.024 |
1.333 ± 0.012 | 2.299 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
