
Lake Sarah-associated circular virus-49
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
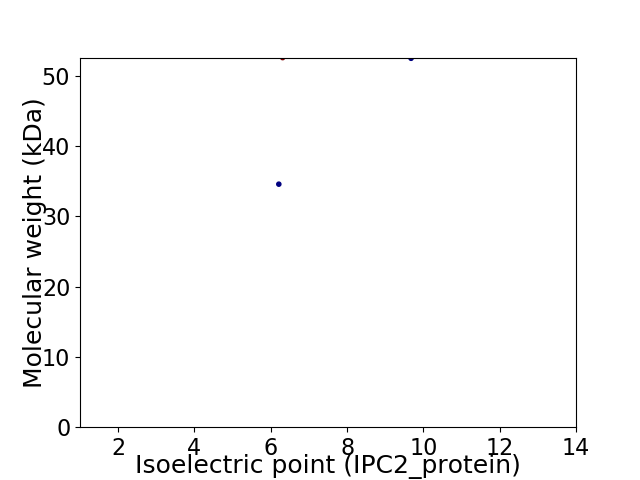
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
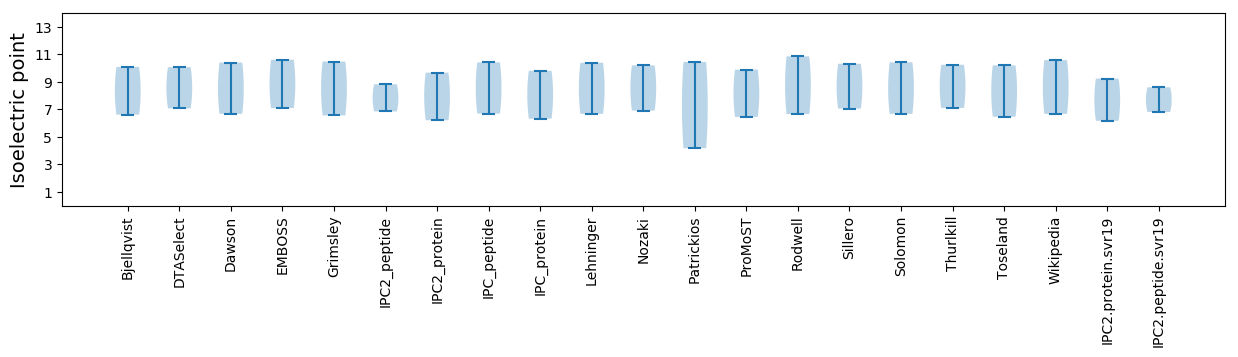
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GAA2|A0A126GAA2_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-49 OX=1685778 PE=4 SV=1
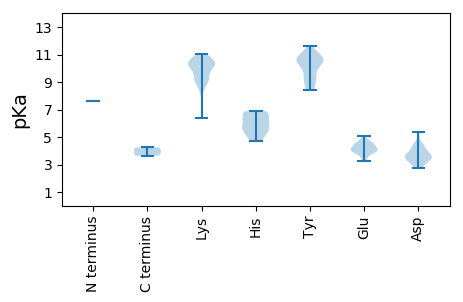
MM1 pKa = 7.6SNAQRR6 pKa = 11.84SGRR9 pKa = 11.84TQGHH13 pKa = 5.65WFIGTCRR20 pKa = 11.84DD21 pKa = 3.64LDD23 pKa = 3.62WDD25 pKa = 4.34GRR27 pKa = 11.84DD28 pKa = 3.19HH29 pKa = 6.78PQFISYY35 pKa = 10.7GKK37 pKa = 9.9GQLEE41 pKa = 4.04LSEE44 pKa = 4.43SGFRR48 pKa = 11.84HH49 pKa = 4.68WQFVVQCKK57 pKa = 10.32DD58 pKa = 2.88STRR61 pKa = 11.84IAKK64 pKa = 9.35LKK66 pKa = 10.2KK67 pKa = 9.23WLPTCHH73 pKa = 6.53WEE75 pKa = 4.09LTRR78 pKa = 11.84SEE80 pKa = 4.23SAIDD84 pKa = 3.89YY85 pKa = 8.62VWKK88 pKa = 10.5EE89 pKa = 3.82EE90 pKa = 3.85TRR92 pKa = 11.84IEE94 pKa = 4.21GSQFEE99 pKa = 4.43FGVRR103 pKa = 11.84RR104 pKa = 11.84LRR106 pKa = 11.84RR107 pKa = 11.84NAATDD112 pKa = 2.92WDD114 pKa = 4.49VIKK117 pKa = 10.85QRR119 pKa = 11.84VLAGSVTSDD128 pKa = 5.34DD129 pKa = 3.45IPADD133 pKa = 3.06IFVRR137 pKa = 11.84YY138 pKa = 9.29YY139 pKa = 10.99SQLKK143 pKa = 9.44YY144 pKa = 10.94NPFYY148 pKa = 10.66YY149 pKa = 10.13RR150 pKa = 11.84SIAKK154 pKa = 10.08DD155 pKa = 3.36YY156 pKa = 10.72SRR158 pKa = 11.84PAAIIRR164 pKa = 11.84SCVVYY169 pKa = 9.98WGPTGTGKK177 pKa = 10.01SRR179 pKa = 11.84RR180 pKa = 11.84AWSEE184 pKa = 3.61ATTAAYY190 pKa = 10.59VKK192 pKa = 10.69DD193 pKa = 4.28PLSKK197 pKa = 9.3WWEE200 pKa = 4.59GYY202 pKa = 9.39QGEE205 pKa = 4.18ISVIIDD211 pKa = 3.4EE212 pKa = 4.68FRR214 pKa = 11.84GIIQPAHH221 pKa = 6.77LLRR224 pKa = 11.84WLDD227 pKa = 3.7RR228 pKa = 11.84YY229 pKa = 9.54PVSVEE234 pKa = 4.09CKK236 pKa = 10.37GGSTPLRR243 pKa = 11.84AQRR246 pKa = 11.84FWICSNLHH254 pKa = 5.62PSQWYY259 pKa = 9.2PEE261 pKa = 4.42LDD263 pKa = 3.3GATYY267 pKa = 10.52SALEE271 pKa = 3.73RR272 pKa = 11.84RR273 pKa = 11.84MEE275 pKa = 4.0IVEE278 pKa = 4.06MAEE281 pKa = 3.38NWEE284 pKa = 4.47PIEE287 pKa = 4.21SEE289 pKa = 3.98EE290 pKa = 4.63LIEE293 pKa = 4.52VSEE296 pKa = 4.24
MM1 pKa = 7.6SNAQRR6 pKa = 11.84SGRR9 pKa = 11.84TQGHH13 pKa = 5.65WFIGTCRR20 pKa = 11.84DD21 pKa = 3.64LDD23 pKa = 3.62WDD25 pKa = 4.34GRR27 pKa = 11.84DD28 pKa = 3.19HH29 pKa = 6.78PQFISYY35 pKa = 10.7GKK37 pKa = 9.9GQLEE41 pKa = 4.04LSEE44 pKa = 4.43SGFRR48 pKa = 11.84HH49 pKa = 4.68WQFVVQCKK57 pKa = 10.32DD58 pKa = 2.88STRR61 pKa = 11.84IAKK64 pKa = 9.35LKK66 pKa = 10.2KK67 pKa = 9.23WLPTCHH73 pKa = 6.53WEE75 pKa = 4.09LTRR78 pKa = 11.84SEE80 pKa = 4.23SAIDD84 pKa = 3.89YY85 pKa = 8.62VWKK88 pKa = 10.5EE89 pKa = 3.82EE90 pKa = 3.85TRR92 pKa = 11.84IEE94 pKa = 4.21GSQFEE99 pKa = 4.43FGVRR103 pKa = 11.84RR104 pKa = 11.84LRR106 pKa = 11.84RR107 pKa = 11.84NAATDD112 pKa = 2.92WDD114 pKa = 4.49VIKK117 pKa = 10.85QRR119 pKa = 11.84VLAGSVTSDD128 pKa = 5.34DD129 pKa = 3.45IPADD133 pKa = 3.06IFVRR137 pKa = 11.84YY138 pKa = 9.29YY139 pKa = 10.99SQLKK143 pKa = 9.44YY144 pKa = 10.94NPFYY148 pKa = 10.66YY149 pKa = 10.13RR150 pKa = 11.84SIAKK154 pKa = 10.08DD155 pKa = 3.36YY156 pKa = 10.72SRR158 pKa = 11.84PAAIIRR164 pKa = 11.84SCVVYY169 pKa = 9.98WGPTGTGKK177 pKa = 10.01SRR179 pKa = 11.84RR180 pKa = 11.84AWSEE184 pKa = 3.61ATTAAYY190 pKa = 10.59VKK192 pKa = 10.69DD193 pKa = 4.28PLSKK197 pKa = 9.3WWEE200 pKa = 4.59GYY202 pKa = 9.39QGEE205 pKa = 4.18ISVIIDD211 pKa = 3.4EE212 pKa = 4.68FRR214 pKa = 11.84GIIQPAHH221 pKa = 6.77LLRR224 pKa = 11.84WLDD227 pKa = 3.7RR228 pKa = 11.84YY229 pKa = 9.54PVSVEE234 pKa = 4.09CKK236 pKa = 10.37GGSTPLRR243 pKa = 11.84AQRR246 pKa = 11.84FWICSNLHH254 pKa = 5.62PSQWYY259 pKa = 9.2PEE261 pKa = 4.42LDD263 pKa = 3.3GATYY267 pKa = 10.52SALEE271 pKa = 3.73RR272 pKa = 11.84RR273 pKa = 11.84MEE275 pKa = 4.0IVEE278 pKa = 4.06MAEE281 pKa = 3.38NWEE284 pKa = 4.47PIEE287 pKa = 4.21SEE289 pKa = 3.98EE290 pKa = 4.63LIEE293 pKa = 4.52VSEE296 pKa = 4.24
Molecular weight: 34.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GAA2|A0A126GAA2_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-49 OX=1685778 PE=4 SV=1
MM1 pKa = 7.61SITRR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.59SAPSVAYY14 pKa = 8.9STAANIANYY23 pKa = 10.01AIRR26 pKa = 11.84HH27 pKa = 5.02PQQVIQGVRR36 pKa = 11.84RR37 pKa = 11.84LSRR40 pKa = 11.84RR41 pKa = 11.84FSEE44 pKa = 5.04AFSTPQGRR52 pKa = 11.84RR53 pKa = 11.84LRR55 pKa = 11.84SNPIPASGSAPGQSKK70 pKa = 10.28RR71 pKa = 11.84YY72 pKa = 9.09AVSQGRR78 pKa = 11.84SFVGASKK85 pKa = 10.95YY86 pKa = 10.36GGSVIVKK93 pKa = 10.31GGFKK97 pKa = 10.18KK98 pKa = 10.6KK99 pKa = 9.09KK100 pKa = 6.36TFKK103 pKa = 9.63RR104 pKa = 11.84KK105 pKa = 8.6SMKK108 pKa = 10.56RR109 pKa = 11.84MMKK112 pKa = 10.19RR113 pKa = 11.84KK114 pKa = 9.13SKK116 pKa = 10.59KK117 pKa = 9.19GTLTDD122 pKa = 4.06CLTKK126 pKa = 10.4GYY128 pKa = 10.35HH129 pKa = 5.29VTDD132 pKa = 3.8QIYY135 pKa = 10.83GLAQDD140 pKa = 4.43PSAVYY145 pKa = 8.45ITHH148 pKa = 6.58SSWHH152 pKa = 5.95LAQMTRR158 pKa = 11.84VIIGAVLRR166 pKa = 11.84KK167 pKa = 9.5ALRR170 pKa = 11.84KK171 pKa = 10.05AGISVPNKK179 pKa = 8.99DD180 pKa = 3.7TLLPLYY186 pKa = 10.71NDD188 pKa = 3.9GNSGRR193 pKa = 11.84MEE195 pKa = 4.0FQYY198 pKa = 10.56TYY200 pKa = 11.21QNPTSGALAAIVTVPTATGSSFDD223 pKa = 3.75TMLGTLMSSGMGTQIQDD240 pKa = 3.59FLSTVSTNQPHH251 pKa = 6.06SFGVYY256 pKa = 9.35TLDD259 pKa = 4.58SGNQTSMMSRR269 pKa = 11.84IYY271 pKa = 10.31LQNEE275 pKa = 3.88YY276 pKa = 10.57FYY278 pKa = 10.73IPVFSSLKK286 pKa = 8.97FQNRR290 pKa = 11.84TLPATGDD297 pKa = 3.26ATVPNIEE304 pKa = 4.52RR305 pKa = 11.84SDD307 pKa = 3.58VQPIKK312 pKa = 11.05VKK314 pKa = 10.45VFHH317 pKa = 6.87FSAGTPRR324 pKa = 11.84LSFVSTTSATPAVTTNVPMGVVNSNGLFLQTALTIGNVDD363 pKa = 3.65YY364 pKa = 11.22QEE366 pKa = 4.83PPTKK370 pKa = 10.4KK371 pKa = 10.18LWTNCISTSNTVVDD385 pKa = 4.18PGIIKK390 pKa = 10.5YY391 pKa = 10.64SSLKK395 pKa = 8.57YY396 pKa = 8.51TYY398 pKa = 10.6KK399 pKa = 10.25GTLSNLQAKK408 pKa = 8.45LRR410 pKa = 11.84PMKK413 pKa = 10.29YY414 pKa = 8.54DD415 pKa = 2.77TGTTVVTGVAGKK427 pKa = 9.1SQIICIDD434 pKa = 3.34EE435 pKa = 4.14YY436 pKa = 11.64LRR438 pKa = 11.84TTTSNKK444 pKa = 9.44IAVSYY449 pKa = 10.77EE450 pKa = 3.28KK451 pKa = 10.34DD452 pKa = 3.46LRR454 pKa = 11.84IGCYY458 pKa = 9.74CVTKK462 pKa = 10.51KK463 pKa = 10.7EE464 pKa = 3.92PAFTSFLNVPAALNIGG480 pKa = 3.64
MM1 pKa = 7.61SITRR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.59SAPSVAYY14 pKa = 8.9STAANIANYY23 pKa = 10.01AIRR26 pKa = 11.84HH27 pKa = 5.02PQQVIQGVRR36 pKa = 11.84RR37 pKa = 11.84LSRR40 pKa = 11.84RR41 pKa = 11.84FSEE44 pKa = 5.04AFSTPQGRR52 pKa = 11.84RR53 pKa = 11.84LRR55 pKa = 11.84SNPIPASGSAPGQSKK70 pKa = 10.28RR71 pKa = 11.84YY72 pKa = 9.09AVSQGRR78 pKa = 11.84SFVGASKK85 pKa = 10.95YY86 pKa = 10.36GGSVIVKK93 pKa = 10.31GGFKK97 pKa = 10.18KK98 pKa = 10.6KK99 pKa = 9.09KK100 pKa = 6.36TFKK103 pKa = 9.63RR104 pKa = 11.84KK105 pKa = 8.6SMKK108 pKa = 10.56RR109 pKa = 11.84MMKK112 pKa = 10.19RR113 pKa = 11.84KK114 pKa = 9.13SKK116 pKa = 10.59KK117 pKa = 9.19GTLTDD122 pKa = 4.06CLTKK126 pKa = 10.4GYY128 pKa = 10.35HH129 pKa = 5.29VTDD132 pKa = 3.8QIYY135 pKa = 10.83GLAQDD140 pKa = 4.43PSAVYY145 pKa = 8.45ITHH148 pKa = 6.58SSWHH152 pKa = 5.95LAQMTRR158 pKa = 11.84VIIGAVLRR166 pKa = 11.84KK167 pKa = 9.5ALRR170 pKa = 11.84KK171 pKa = 10.05AGISVPNKK179 pKa = 8.99DD180 pKa = 3.7TLLPLYY186 pKa = 10.71NDD188 pKa = 3.9GNSGRR193 pKa = 11.84MEE195 pKa = 4.0FQYY198 pKa = 10.56TYY200 pKa = 11.21QNPTSGALAAIVTVPTATGSSFDD223 pKa = 3.75TMLGTLMSSGMGTQIQDD240 pKa = 3.59FLSTVSTNQPHH251 pKa = 6.06SFGVYY256 pKa = 9.35TLDD259 pKa = 4.58SGNQTSMMSRR269 pKa = 11.84IYY271 pKa = 10.31LQNEE275 pKa = 3.88YY276 pKa = 10.57FYY278 pKa = 10.73IPVFSSLKK286 pKa = 8.97FQNRR290 pKa = 11.84TLPATGDD297 pKa = 3.26ATVPNIEE304 pKa = 4.52RR305 pKa = 11.84SDD307 pKa = 3.58VQPIKK312 pKa = 11.05VKK314 pKa = 10.45VFHH317 pKa = 6.87FSAGTPRR324 pKa = 11.84LSFVSTTSATPAVTTNVPMGVVNSNGLFLQTALTIGNVDD363 pKa = 3.65YY364 pKa = 11.22QEE366 pKa = 4.83PPTKK370 pKa = 10.4KK371 pKa = 10.18LWTNCISTSNTVVDD385 pKa = 4.18PGIIKK390 pKa = 10.5YY391 pKa = 10.64SSLKK395 pKa = 8.57YY396 pKa = 8.51TYY398 pKa = 10.6KK399 pKa = 10.25GTLSNLQAKK408 pKa = 8.45LRR410 pKa = 11.84PMKK413 pKa = 10.29YY414 pKa = 8.54DD415 pKa = 2.77TGTTVVTGVAGKK427 pKa = 9.1SQIICIDD434 pKa = 3.34EE435 pKa = 4.14YY436 pKa = 11.64LRR438 pKa = 11.84TTTSNKK444 pKa = 9.44IAVSYY449 pKa = 10.77EE450 pKa = 3.28KK451 pKa = 10.34DD452 pKa = 3.46LRR454 pKa = 11.84IGCYY458 pKa = 9.74CVTKK462 pKa = 10.51KK463 pKa = 10.7EE464 pKa = 3.92PAFTSFLNVPAALNIGG480 pKa = 3.64
Molecular weight: 52.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
776 |
296 |
480 |
388.0 |
43.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.701 ± 0.173 | 1.418 ± 0.374 |
3.995 ± 0.867 | 4.124 ± 2.448 |
3.608 ± 0.141 | 7.088 ± 0.411 |
1.675 ± 0.216 | 6.057 ± 0.43 |
5.928 ± 0.944 | 6.443 ± 0.223 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.062 ± 0.644 | 3.479 ± 1.1 |
4.897 ± 0.31 | 4.51 ± 0.073 |
6.701 ± 1.28 | 9.794 ± 0.621 |
8.119 ± 2.082 | 6.572 ± 0.717 |
2.191 ± 1.767 | 4.639 ± 0.056 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
