
Circovirus-like genome SAR-A
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
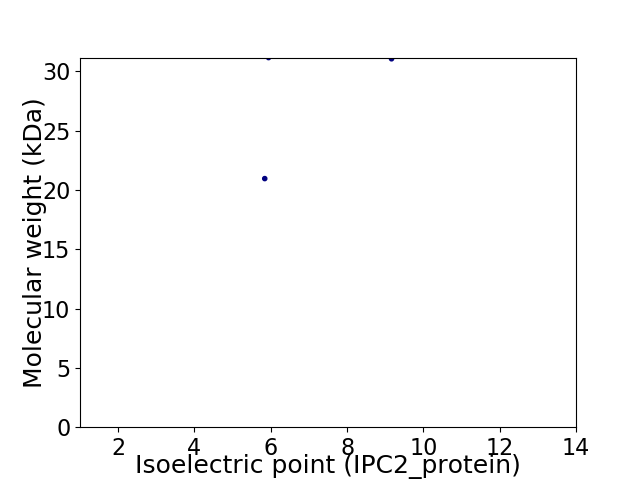
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
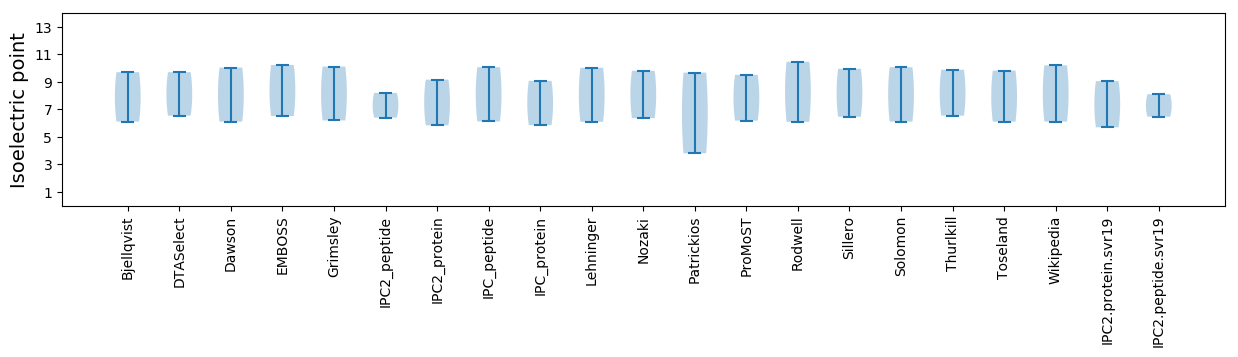
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GIJ1|C6GIJ1_9VIRU Putative Rep protein OS=Circovirus-like genome SAR-A OX=642258 PE=4 SV=2
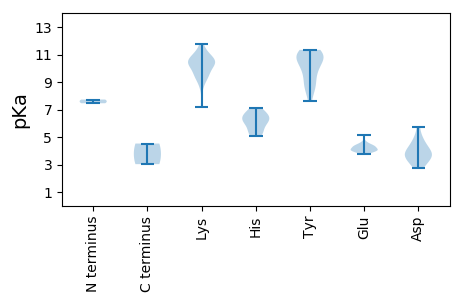
MM1 pKa = 7.46PRR3 pKa = 11.84VNPAKK8 pKa = 10.21AWCFTLNNYY17 pKa = 7.62TEE19 pKa = 4.39NEE21 pKa = 3.98HH22 pKa = 6.14GALVQRR28 pKa = 11.84FSDD31 pKa = 4.35FDD33 pKa = 3.76DD34 pKa = 3.77KK35 pKa = 11.73YY36 pKa = 10.63YY37 pKa = 10.92FIVGCEE43 pKa = 3.74IGAQGTPHH51 pKa = 5.88LQGYY55 pKa = 8.38IEE57 pKa = 4.28KK58 pKa = 10.29KK59 pKa = 9.35VGRR62 pKa = 11.84FRR64 pKa = 11.84PLPCFEE70 pKa = 4.26VLRR73 pKa = 11.84DD74 pKa = 3.94GKK76 pKa = 10.51NAMHH80 pKa = 7.1FEE82 pKa = 4.1RR83 pKa = 11.84AKK85 pKa = 10.98GNRR88 pKa = 11.84KK89 pKa = 8.59QNYY92 pKa = 8.63NYY94 pKa = 9.91CSKK97 pKa = 11.13DD98 pKa = 2.75GDD100 pKa = 4.4FITNIDD106 pKa = 4.28KK107 pKa = 10.82PIMTYY112 pKa = 10.47SEE114 pKa = 5.15AKK116 pKa = 10.45DD117 pKa = 2.88IWKK120 pKa = 7.2EE121 pKa = 4.0TNGISNDD128 pKa = 3.39KK129 pKa = 10.58YY130 pKa = 11.09DD131 pKa = 3.5AATINEE137 pKa = 4.02AAMHH141 pKa = 6.35IEE143 pKa = 4.32YY144 pKa = 9.65MDD146 pKa = 5.72LYY148 pKa = 11.22DD149 pKa = 5.0CYY151 pKa = 10.82TDD153 pKa = 3.8EE154 pKa = 4.41GQKK157 pKa = 10.96KK158 pKa = 9.86FMVRR162 pKa = 11.84YY163 pKa = 8.79KK164 pKa = 11.52AMMQARR170 pKa = 11.84YY171 pKa = 8.83PEE173 pKa = 4.56KK174 pKa = 9.66EE175 pKa = 4.13TVEE178 pKa = 4.04II179 pKa = 4.53
MM1 pKa = 7.46PRR3 pKa = 11.84VNPAKK8 pKa = 10.21AWCFTLNNYY17 pKa = 7.62TEE19 pKa = 4.39NEE21 pKa = 3.98HH22 pKa = 6.14GALVQRR28 pKa = 11.84FSDD31 pKa = 4.35FDD33 pKa = 3.76DD34 pKa = 3.77KK35 pKa = 11.73YY36 pKa = 10.63YY37 pKa = 10.92FIVGCEE43 pKa = 3.74IGAQGTPHH51 pKa = 5.88LQGYY55 pKa = 8.38IEE57 pKa = 4.28KK58 pKa = 10.29KK59 pKa = 9.35VGRR62 pKa = 11.84FRR64 pKa = 11.84PLPCFEE70 pKa = 4.26VLRR73 pKa = 11.84DD74 pKa = 3.94GKK76 pKa = 10.51NAMHH80 pKa = 7.1FEE82 pKa = 4.1RR83 pKa = 11.84AKK85 pKa = 10.98GNRR88 pKa = 11.84KK89 pKa = 8.59QNYY92 pKa = 8.63NYY94 pKa = 9.91CSKK97 pKa = 11.13DD98 pKa = 2.75GDD100 pKa = 4.4FITNIDD106 pKa = 4.28KK107 pKa = 10.82PIMTYY112 pKa = 10.47SEE114 pKa = 5.15AKK116 pKa = 10.45DD117 pKa = 2.88IWKK120 pKa = 7.2EE121 pKa = 4.0TNGISNDD128 pKa = 3.39KK129 pKa = 10.58YY130 pKa = 11.09DD131 pKa = 3.5AATINEE137 pKa = 4.02AAMHH141 pKa = 6.35IEE143 pKa = 4.32YY144 pKa = 9.65MDD146 pKa = 5.72LYY148 pKa = 11.22DD149 pKa = 5.0CYY151 pKa = 10.82TDD153 pKa = 3.8EE154 pKa = 4.41GQKK157 pKa = 10.96KK158 pKa = 9.86FMVRR162 pKa = 11.84YY163 pKa = 8.79KK164 pKa = 11.52AMMQARR170 pKa = 11.84YY171 pKa = 8.83PEE173 pKa = 4.56KK174 pKa = 9.66EE175 pKa = 4.13TVEE178 pKa = 4.04II179 pKa = 4.53
Molecular weight: 20.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GIJ1|C6GIJ1_9VIRU Putative Rep protein OS=Circovirus-like genome SAR-A OX=642258 PE=4 SV=2
MM1 pKa = 7.73PGYY4 pKa = 10.46NFRR7 pKa = 11.84SNPVKK12 pKa = 10.39KK13 pKa = 9.53PSYY16 pKa = 9.92RR17 pKa = 11.84FRR19 pKa = 11.84SKK21 pKa = 10.67SKK23 pKa = 10.17PRR25 pKa = 11.84SQPNKK30 pKa = 9.12SVKK33 pKa = 10.46KK34 pKa = 9.54EE35 pKa = 4.0VKK37 pKa = 10.47KK38 pKa = 9.42NTKK41 pKa = 9.7DD42 pKa = 3.1IKK44 pKa = 10.45LLKK47 pKa = 10.61SVGFQYY53 pKa = 11.34APFQQRR59 pKa = 11.84EE60 pKa = 4.05VGTISNFLHH69 pKa = 6.39TSLLTAPNNWGGIFRR84 pKa = 11.84MHH86 pKa = 6.53GVSNEE91 pKa = 3.89DD92 pKa = 3.5LPRR95 pKa = 11.84QYY97 pKa = 10.77MMKK100 pKa = 10.19SVDD103 pKa = 3.87VNWIAQAEE111 pKa = 4.02ASEE114 pKa = 4.64VGNLWLQVFVVSLKK128 pKa = 10.85HH129 pKa = 6.63KK130 pKa = 8.95MANQVLTRR138 pKa = 11.84TTRR141 pKa = 11.84LTNLEE146 pKa = 4.16EE147 pKa = 4.36GLDD150 pKa = 3.92YY151 pKa = 10.9TSQSAGTAFALQGDD165 pKa = 4.94LGYY168 pKa = 11.29KK169 pKa = 10.28LNPDD173 pKa = 4.51LYY175 pKa = 9.23TIHH178 pKa = 6.05YY179 pKa = 9.92HH180 pKa = 5.45SGQRR184 pKa = 11.84RR185 pKa = 11.84IGEE188 pKa = 4.4STVGEE193 pKa = 4.33SPVAVTNINNGTTMGSCRR211 pKa = 11.84IKK213 pKa = 10.23WPHH216 pKa = 5.1TFKK219 pKa = 11.07NDD221 pKa = 3.16EE222 pKa = 4.09TSDD225 pKa = 3.47AGFKK229 pKa = 10.49EE230 pKa = 3.87LTYY233 pKa = 11.16QNIEE237 pKa = 4.32PKK239 pKa = 9.51QHH241 pKa = 6.84LYY243 pKa = 10.79LLVMSNTSGSVITGGEE259 pKa = 4.06LFFSSRR265 pKa = 11.84AQFNGHH271 pKa = 5.29VNNPNN276 pKa = 3.03
MM1 pKa = 7.73PGYY4 pKa = 10.46NFRR7 pKa = 11.84SNPVKK12 pKa = 10.39KK13 pKa = 9.53PSYY16 pKa = 9.92RR17 pKa = 11.84FRR19 pKa = 11.84SKK21 pKa = 10.67SKK23 pKa = 10.17PRR25 pKa = 11.84SQPNKK30 pKa = 9.12SVKK33 pKa = 10.46KK34 pKa = 9.54EE35 pKa = 4.0VKK37 pKa = 10.47KK38 pKa = 9.42NTKK41 pKa = 9.7DD42 pKa = 3.1IKK44 pKa = 10.45LLKK47 pKa = 10.61SVGFQYY53 pKa = 11.34APFQQRR59 pKa = 11.84EE60 pKa = 4.05VGTISNFLHH69 pKa = 6.39TSLLTAPNNWGGIFRR84 pKa = 11.84MHH86 pKa = 6.53GVSNEE91 pKa = 3.89DD92 pKa = 3.5LPRR95 pKa = 11.84QYY97 pKa = 10.77MMKK100 pKa = 10.19SVDD103 pKa = 3.87VNWIAQAEE111 pKa = 4.02ASEE114 pKa = 4.64VGNLWLQVFVVSLKK128 pKa = 10.85HH129 pKa = 6.63KK130 pKa = 8.95MANQVLTRR138 pKa = 11.84TTRR141 pKa = 11.84LTNLEE146 pKa = 4.16EE147 pKa = 4.36GLDD150 pKa = 3.92YY151 pKa = 10.9TSQSAGTAFALQGDD165 pKa = 4.94LGYY168 pKa = 11.29KK169 pKa = 10.28LNPDD173 pKa = 4.51LYY175 pKa = 9.23TIHH178 pKa = 6.05YY179 pKa = 9.92HH180 pKa = 5.45SGQRR184 pKa = 11.84RR185 pKa = 11.84IGEE188 pKa = 4.4STVGEE193 pKa = 4.33SPVAVTNINNGTTMGSCRR211 pKa = 11.84IKK213 pKa = 10.23WPHH216 pKa = 5.1TFKK219 pKa = 11.07NDD221 pKa = 3.16EE222 pKa = 4.09TSDD225 pKa = 3.47AGFKK229 pKa = 10.49EE230 pKa = 3.87LTYY233 pKa = 11.16QNIEE237 pKa = 4.32PKK239 pKa = 9.51QHH241 pKa = 6.84LYY243 pKa = 10.79LLVMSNTSGSVITGGEE259 pKa = 4.06LFFSSRR265 pKa = 11.84AQFNGHH271 pKa = 5.29VNNPNN276 pKa = 3.03
Molecular weight: 31.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
455 |
179 |
276 |
227.5 |
26.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.495 ± 1.092 | 1.319 ± 0.911 |
4.615 ± 1.635 | 5.934 ± 1.166 |
4.835 ± 0.119 | 7.033 ± 0.548 |
2.637 ± 0.249 | 4.615 ± 0.945 |
7.912 ± 0.634 | 6.154 ± 1.731 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.297 ± 0.724 | 7.692 ± 0.611 |
4.396 ± 0.3 | 4.396 ± 0.645 |
4.835 ± 0.119 | 6.374 ± 2.557 |
6.374 ± 0.831 | 5.714 ± 1.114 |
1.319 ± 0.124 | 5.055 ± 1.364 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
