
Robbsia andropogonis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Robbsia
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
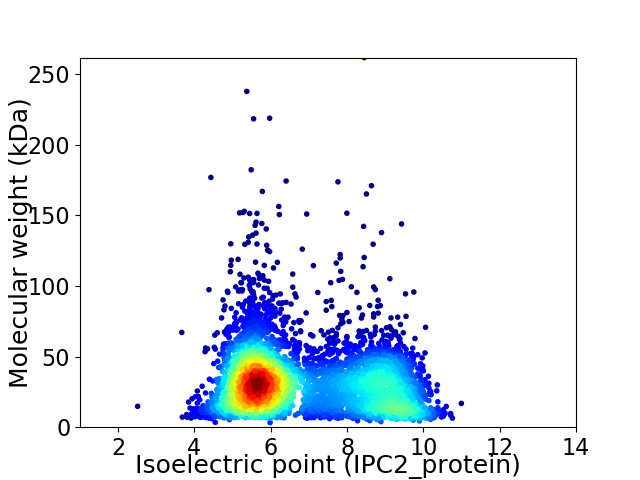
Virtual 2D-PAGE plot for 4737 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F5JTQ6|A0A0F5JTQ6_9BURK Uncharacterized protein (Fragment) OS=Robbsia andropogonis OX=28092 GN=WM40_27485 PE=4 SV=1
MM1 pKa = 7.72ILDD4 pKa = 4.13GKK6 pKa = 10.38GGNDD10 pKa = 3.53TYY12 pKa = 11.27HH13 pKa = 6.96SYY15 pKa = 10.53YY16 pKa = 11.31ANDD19 pKa = 3.41TFVFNSGYY27 pKa = 10.38GALEE31 pKa = 3.97INNEE35 pKa = 4.09NYY37 pKa = 10.55SGGTFASVLRR47 pKa = 11.84FGPGVTLDD55 pKa = 3.78SLRR58 pKa = 11.84VTSDD62 pKa = 3.01GNVLVLRR69 pKa = 11.84DD70 pKa = 4.1GVDD73 pKa = 3.46GDD75 pKa = 4.54AVTVDD80 pKa = 3.6HH81 pKa = 6.75FFSEE85 pKa = 4.7FGWAGITSVEE95 pKa = 4.22LNDD98 pKa = 3.7GTVLNVSQLIQLEE111 pKa = 4.59EE112 pKa = 4.15TGSTGADD119 pKa = 2.98TLYY122 pKa = 9.58GTSGADD128 pKa = 3.42TIDD131 pKa = 3.38GRR133 pKa = 11.84GGNDD137 pKa = 3.22LAIGNGGADD146 pKa = 3.21TFFYY150 pKa = 10.45NAGYY154 pKa = 9.58GALEE158 pKa = 4.08ISSDD162 pKa = 3.7EE163 pKa = 4.27GSTPTSVLRR172 pKa = 11.84FGEE175 pKa = 4.99GITASSITVRR185 pKa = 11.84NSNSGTAIQITDD197 pKa = 4.84GIDD200 pKa = 3.34GDD202 pKa = 4.78VITIDD207 pKa = 4.1NMYY210 pKa = 10.3SDD212 pKa = 4.91SGTKK216 pKa = 10.36GVGSVEE222 pKa = 4.18FFDD225 pKa = 4.29GTTLTAQQLIALNAGRR241 pKa = 11.84APEE244 pKa = 3.8ATYY247 pKa = 11.12YY248 pKa = 9.2GTTGADD254 pKa = 3.57SITGSGEE261 pKa = 4.15DD262 pKa = 3.87EE263 pKa = 4.63LFDD266 pKa = 4.28GKK268 pKa = 11.24GGTDD272 pKa = 3.48YY273 pKa = 11.41FKK275 pKa = 11.58GNAGNDD281 pKa = 3.19TFVFNQGYY289 pKa = 8.53RR290 pKa = 11.84ALEE293 pKa = 3.89VDD295 pKa = 4.09EE296 pKa = 4.58NWYY299 pKa = 10.58SGQAPVLQLGAGIEE313 pKa = 3.97ASMLKK318 pKa = 10.48VSVANSHH325 pKa = 6.26SGLVITDD332 pKa = 3.9GVAGDD337 pKa = 5.14QITLDD342 pKa = 3.47NMLYY346 pKa = 10.25DD347 pKa = 3.95YY348 pKa = 11.2QGVSTIRR355 pKa = 11.84FADD358 pKa = 4.05GSTLSKK364 pKa = 10.7AQIIAMEE371 pKa = 4.31TTGTSGADD379 pKa = 3.06SMYY382 pKa = 10.84GSTAAEE388 pKa = 4.13LFDD391 pKa = 4.18GKK393 pKa = 11.04GGADD397 pKa = 3.49YY398 pKa = 11.41AKK400 pKa = 10.73GSGGNDD406 pKa = 2.49TFVFNEE412 pKa = 5.19GYY414 pKa = 9.88GQLEE418 pKa = 3.76IDD420 pKa = 3.46EE421 pKa = 4.65TLNDD425 pKa = 3.65GATTVLQLGAGITRR439 pKa = 11.84EE440 pKa = 3.96NIKK443 pKa = 10.7AYY445 pKa = 10.42FDD447 pKa = 3.72GATLVLTDD455 pKa = 5.65GISGDD460 pKa = 3.83QIRR463 pKa = 11.84IDD465 pKa = 3.45NEE467 pKa = 4.07KK468 pKa = 10.73YY469 pKa = 10.51SNNGINLVQFADD481 pKa = 4.21GATLTQADD489 pKa = 4.91LQTLPTTGSASNDD502 pKa = 3.42SLTGTGDD509 pKa = 3.49SEE511 pKa = 4.93VIDD514 pKa = 4.0GKK516 pKa = 11.22GGNDD520 pKa = 3.18TVNGNMGNDD529 pKa = 3.12TFVYY533 pKa = 8.58NQGYY537 pKa = 8.28GALEE541 pKa = 3.77INNNYY546 pKa = 9.55WYY548 pKa = 10.11GQNPVLQLGAGITAADD564 pKa = 3.91LQVATDD570 pKa = 3.84ASHH573 pKa = 6.81TGLILTDD580 pKa = 4.13GVAGDD585 pKa = 4.89TIHH588 pKa = 7.33IDD590 pKa = 3.6NIKK593 pKa = 8.51STEE596 pKa = 3.96RR597 pKa = 11.84TGVGSVTFADD607 pKa = 4.31GTTMSAADD615 pKa = 5.24LIALTTVGTTGNDD628 pKa = 3.19ALYY631 pKa = 10.49GSSSNDD637 pKa = 3.05MFDD640 pKa = 4.14GKK642 pKa = 11.1GGDD645 pKa = 3.72DD646 pKa = 4.03TITT649 pKa = 3.17
MM1 pKa = 7.72ILDD4 pKa = 4.13GKK6 pKa = 10.38GGNDD10 pKa = 3.53TYY12 pKa = 11.27HH13 pKa = 6.96SYY15 pKa = 10.53YY16 pKa = 11.31ANDD19 pKa = 3.41TFVFNSGYY27 pKa = 10.38GALEE31 pKa = 3.97INNEE35 pKa = 4.09NYY37 pKa = 10.55SGGTFASVLRR47 pKa = 11.84FGPGVTLDD55 pKa = 3.78SLRR58 pKa = 11.84VTSDD62 pKa = 3.01GNVLVLRR69 pKa = 11.84DD70 pKa = 4.1GVDD73 pKa = 3.46GDD75 pKa = 4.54AVTVDD80 pKa = 3.6HH81 pKa = 6.75FFSEE85 pKa = 4.7FGWAGITSVEE95 pKa = 4.22LNDD98 pKa = 3.7GTVLNVSQLIQLEE111 pKa = 4.59EE112 pKa = 4.15TGSTGADD119 pKa = 2.98TLYY122 pKa = 9.58GTSGADD128 pKa = 3.42TIDD131 pKa = 3.38GRR133 pKa = 11.84GGNDD137 pKa = 3.22LAIGNGGADD146 pKa = 3.21TFFYY150 pKa = 10.45NAGYY154 pKa = 9.58GALEE158 pKa = 4.08ISSDD162 pKa = 3.7EE163 pKa = 4.27GSTPTSVLRR172 pKa = 11.84FGEE175 pKa = 4.99GITASSITVRR185 pKa = 11.84NSNSGTAIQITDD197 pKa = 4.84GIDD200 pKa = 3.34GDD202 pKa = 4.78VITIDD207 pKa = 4.1NMYY210 pKa = 10.3SDD212 pKa = 4.91SGTKK216 pKa = 10.36GVGSVEE222 pKa = 4.18FFDD225 pKa = 4.29GTTLTAQQLIALNAGRR241 pKa = 11.84APEE244 pKa = 3.8ATYY247 pKa = 11.12YY248 pKa = 9.2GTTGADD254 pKa = 3.57SITGSGEE261 pKa = 4.15DD262 pKa = 3.87EE263 pKa = 4.63LFDD266 pKa = 4.28GKK268 pKa = 11.24GGTDD272 pKa = 3.48YY273 pKa = 11.41FKK275 pKa = 11.58GNAGNDD281 pKa = 3.19TFVFNQGYY289 pKa = 8.53RR290 pKa = 11.84ALEE293 pKa = 3.89VDD295 pKa = 4.09EE296 pKa = 4.58NWYY299 pKa = 10.58SGQAPVLQLGAGIEE313 pKa = 3.97ASMLKK318 pKa = 10.48VSVANSHH325 pKa = 6.26SGLVITDD332 pKa = 3.9GVAGDD337 pKa = 5.14QITLDD342 pKa = 3.47NMLYY346 pKa = 10.25DD347 pKa = 3.95YY348 pKa = 11.2QGVSTIRR355 pKa = 11.84FADD358 pKa = 4.05GSTLSKK364 pKa = 10.7AQIIAMEE371 pKa = 4.31TTGTSGADD379 pKa = 3.06SMYY382 pKa = 10.84GSTAAEE388 pKa = 4.13LFDD391 pKa = 4.18GKK393 pKa = 11.04GGADD397 pKa = 3.49YY398 pKa = 11.41AKK400 pKa = 10.73GSGGNDD406 pKa = 2.49TFVFNEE412 pKa = 5.19GYY414 pKa = 9.88GQLEE418 pKa = 3.76IDD420 pKa = 3.46EE421 pKa = 4.65TLNDD425 pKa = 3.65GATTVLQLGAGITRR439 pKa = 11.84EE440 pKa = 3.96NIKK443 pKa = 10.7AYY445 pKa = 10.42FDD447 pKa = 3.72GATLVLTDD455 pKa = 5.65GISGDD460 pKa = 3.83QIRR463 pKa = 11.84IDD465 pKa = 3.45NEE467 pKa = 4.07KK468 pKa = 10.73YY469 pKa = 10.51SNNGINLVQFADD481 pKa = 4.21GATLTQADD489 pKa = 4.91LQTLPTTGSASNDD502 pKa = 3.42SLTGTGDD509 pKa = 3.49SEE511 pKa = 4.93VIDD514 pKa = 4.0GKK516 pKa = 11.22GGNDD520 pKa = 3.18TVNGNMGNDD529 pKa = 3.12TFVYY533 pKa = 8.58NQGYY537 pKa = 8.28GALEE541 pKa = 3.77INNNYY546 pKa = 9.55WYY548 pKa = 10.11GQNPVLQLGAGITAADD564 pKa = 3.91LQVATDD570 pKa = 3.84ASHH573 pKa = 6.81TGLILTDD580 pKa = 4.13GVAGDD585 pKa = 4.89TIHH588 pKa = 7.33IDD590 pKa = 3.6NIKK593 pKa = 8.51STEE596 pKa = 3.96RR597 pKa = 11.84TGVGSVTFADD607 pKa = 4.31GTTMSAADD615 pKa = 5.24LIALTTVGTTGNDD628 pKa = 3.19ALYY631 pKa = 10.49GSSSNDD637 pKa = 3.05MFDD640 pKa = 4.14GKK642 pKa = 11.1GGDD645 pKa = 3.72DD646 pKa = 4.03TITT649 pKa = 3.17
Molecular weight: 67.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F5JTU6|A0A0F5JTU6_9BURK Uncharacterized protein OS=Robbsia andropogonis OX=28092 GN=WM40_25385 PE=4 SV=1
MM1 pKa = 7.56AGWSLAWEE9 pKa = 4.43EE10 pKa = 5.34DD11 pKa = 3.51NLLHH15 pKa = 6.61CKK17 pKa = 10.35SFSHH21 pKa = 6.7CEE23 pKa = 3.54KK24 pKa = 10.33RR25 pKa = 11.84SRR27 pKa = 11.84NLKK30 pKa = 9.92IVVALSAGASYY41 pKa = 11.02NPHH44 pKa = 6.57RR45 pKa = 11.84LSAFRR50 pKa = 11.84AGARR54 pKa = 11.84VVRR57 pKa = 11.84IGMAYY62 pKa = 10.04AVASVCRR69 pKa = 11.84VAGSVLRR76 pKa = 11.84PVGRR80 pKa = 11.84DD81 pKa = 3.04AILRR85 pKa = 11.84RR86 pKa = 11.84QPCPQCQRR94 pKa = 11.84KK95 pKa = 8.69
MM1 pKa = 7.56AGWSLAWEE9 pKa = 4.43EE10 pKa = 5.34DD11 pKa = 3.51NLLHH15 pKa = 6.61CKK17 pKa = 10.35SFSHH21 pKa = 6.7CEE23 pKa = 3.54KK24 pKa = 10.33RR25 pKa = 11.84SRR27 pKa = 11.84NLKK30 pKa = 9.92IVVALSAGASYY41 pKa = 11.02NPHH44 pKa = 6.57RR45 pKa = 11.84LSAFRR50 pKa = 11.84AGARR54 pKa = 11.84VVRR57 pKa = 11.84IGMAYY62 pKa = 10.04AVASVCRR69 pKa = 11.84VAGSVLRR76 pKa = 11.84PVGRR80 pKa = 11.84DD81 pKa = 3.04AILRR85 pKa = 11.84RR86 pKa = 11.84QPCPQCQRR94 pKa = 11.84KK95 pKa = 8.69
Molecular weight: 10.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1470760 |
29 |
2373 |
310.5 |
33.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.419 ± 0.051 | 0.944 ± 0.01 |
5.683 ± 0.029 | 4.697 ± 0.03 |
3.598 ± 0.024 | 7.99 ± 0.031 |
2.419 ± 0.018 | 4.997 ± 0.027 |
3.027 ± 0.031 | 10.122 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.522 ± 0.016 | 2.845 ± 0.024 |
5.051 ± 0.027 | 3.653 ± 0.02 |
7.025 ± 0.039 | 5.96 ± 0.032 |
5.696 ± 0.029 | 7.565 ± 0.028 |
1.391 ± 0.013 | 2.396 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
