
Dragonfly circularisvirus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Circularisvirus
Average proteome isoelectric point is 8.36
Get precalculated fractions of proteins
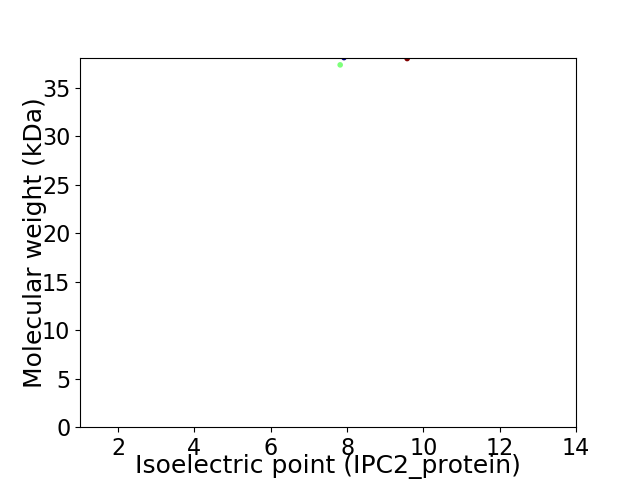
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0A2Q4|K0A2Q4_9VIRU Replication-associated protein OS=Dragonfly circularisvirus OX=1234872 PE=4 SV=1
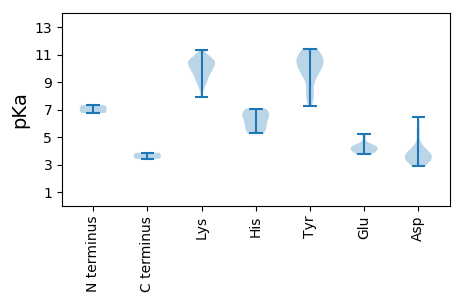
MM1 pKa = 7.32SRR3 pKa = 11.84CKK5 pKa = 10.06NWCFTSFNVDD15 pKa = 3.43SPPKK19 pKa = 9.57FDD21 pKa = 6.43PITTEE26 pKa = 3.74YY27 pKa = 10.66LCYY30 pKa = 10.48GRR32 pKa = 11.84EE33 pKa = 4.03TCPTTKK39 pKa = 9.99KK40 pKa = 10.25SHH42 pKa = 5.73LQGFICLKK50 pKa = 8.64EE51 pKa = 4.34RR52 pKa = 11.84KK53 pKa = 9.25RR54 pKa = 11.84LAGVKK59 pKa = 9.79KK60 pKa = 10.64LLGSVHH66 pKa = 7.07LEE68 pKa = 3.87ASRR71 pKa = 11.84GTVDD75 pKa = 5.28QNIQYY80 pKa = 9.95CSKK83 pKa = 10.72DD84 pKa = 3.37GEE86 pKa = 4.3FTEE89 pKa = 5.21FGSRR93 pKa = 11.84PSSGPATSPFALAIAAATDD112 pKa = 3.24GRR114 pKa = 11.84LDD116 pKa = 3.97DD117 pKa = 4.6VKK119 pKa = 10.42STHH122 pKa = 6.33PGIYY126 pKa = 9.97LRR128 pKa = 11.84YY129 pKa = 9.29KK130 pKa = 9.28KK131 pKa = 8.89TLEE134 pKa = 3.86SLQKK138 pKa = 10.33FRR140 pKa = 11.84SDD142 pKa = 5.42DD143 pKa = 3.75LDD145 pKa = 4.03GSCGVWICGPPRR157 pKa = 11.84IGKK160 pKa = 9.76DD161 pKa = 3.2YY162 pKa = 11.1AVRR165 pKa = 11.84KK166 pKa = 9.24IGNVYY171 pKa = 10.38CKK173 pKa = 10.45ALNKK177 pKa = 8.98WWDD180 pKa = 3.78GYY182 pKa = 11.03LGEE185 pKa = 4.7PNVLISDD192 pKa = 4.08VEE194 pKa = 3.97PDD196 pKa = 3.05HH197 pKa = 6.64FKK199 pKa = 10.46WIGYY203 pKa = 7.28FLKK206 pKa = 9.94IWSDD210 pKa = 3.86RR211 pKa = 11.84YY212 pKa = 10.36PFIAEE217 pKa = 3.91IKK219 pKa = 8.93GSSMKK224 pKa = 9.89IRR226 pKa = 11.84PEE228 pKa = 4.19KK229 pKa = 10.72IFVTSNFRR237 pKa = 11.84LSDD240 pKa = 3.7CCNGEE245 pKa = 3.82ILGALQSRR253 pKa = 11.84FTIYY257 pKa = 11.43DD258 pKa = 3.1MFDD261 pKa = 2.86NVVRR265 pKa = 11.84KK266 pKa = 9.8RR267 pKa = 11.84PVVAASTCVFDD278 pKa = 5.9LLVEE282 pKa = 4.46NGDD285 pKa = 3.46ACLEE289 pKa = 4.23KK290 pKa = 10.98EE291 pKa = 4.4NVPPAVQEE299 pKa = 4.26AVATTSKK306 pKa = 10.54QPTRR310 pKa = 11.84AEE312 pKa = 4.05EE313 pKa = 4.26EE314 pKa = 4.15EE315 pKa = 4.27LQDD318 pKa = 3.67FQVVKK323 pKa = 10.66KK324 pKa = 10.18KK325 pKa = 9.94PRR327 pKa = 11.84KK328 pKa = 9.45AKK330 pKa = 10.56SSS332 pKa = 3.41
MM1 pKa = 7.32SRR3 pKa = 11.84CKK5 pKa = 10.06NWCFTSFNVDD15 pKa = 3.43SPPKK19 pKa = 9.57FDD21 pKa = 6.43PITTEE26 pKa = 3.74YY27 pKa = 10.66LCYY30 pKa = 10.48GRR32 pKa = 11.84EE33 pKa = 4.03TCPTTKK39 pKa = 9.99KK40 pKa = 10.25SHH42 pKa = 5.73LQGFICLKK50 pKa = 8.64EE51 pKa = 4.34RR52 pKa = 11.84KK53 pKa = 9.25RR54 pKa = 11.84LAGVKK59 pKa = 9.79KK60 pKa = 10.64LLGSVHH66 pKa = 7.07LEE68 pKa = 3.87ASRR71 pKa = 11.84GTVDD75 pKa = 5.28QNIQYY80 pKa = 9.95CSKK83 pKa = 10.72DD84 pKa = 3.37GEE86 pKa = 4.3FTEE89 pKa = 5.21FGSRR93 pKa = 11.84PSSGPATSPFALAIAAATDD112 pKa = 3.24GRR114 pKa = 11.84LDD116 pKa = 3.97DD117 pKa = 4.6VKK119 pKa = 10.42STHH122 pKa = 6.33PGIYY126 pKa = 9.97LRR128 pKa = 11.84YY129 pKa = 9.29KK130 pKa = 9.28KK131 pKa = 8.89TLEE134 pKa = 3.86SLQKK138 pKa = 10.33FRR140 pKa = 11.84SDD142 pKa = 5.42DD143 pKa = 3.75LDD145 pKa = 4.03GSCGVWICGPPRR157 pKa = 11.84IGKK160 pKa = 9.76DD161 pKa = 3.2YY162 pKa = 11.1AVRR165 pKa = 11.84KK166 pKa = 9.24IGNVYY171 pKa = 10.38CKK173 pKa = 10.45ALNKK177 pKa = 8.98WWDD180 pKa = 3.78GYY182 pKa = 11.03LGEE185 pKa = 4.7PNVLISDD192 pKa = 4.08VEE194 pKa = 3.97PDD196 pKa = 3.05HH197 pKa = 6.64FKK199 pKa = 10.46WIGYY203 pKa = 7.28FLKK206 pKa = 9.94IWSDD210 pKa = 3.86RR211 pKa = 11.84YY212 pKa = 10.36PFIAEE217 pKa = 3.91IKK219 pKa = 8.93GSSMKK224 pKa = 9.89IRR226 pKa = 11.84PEE228 pKa = 4.19KK229 pKa = 10.72IFVTSNFRR237 pKa = 11.84LSDD240 pKa = 3.7CCNGEE245 pKa = 3.82ILGALQSRR253 pKa = 11.84FTIYY257 pKa = 11.43DD258 pKa = 3.1MFDD261 pKa = 2.86NVVRR265 pKa = 11.84KK266 pKa = 9.8RR267 pKa = 11.84PVVAASTCVFDD278 pKa = 5.9LLVEE282 pKa = 4.46NGDD285 pKa = 3.46ACLEE289 pKa = 4.23KK290 pKa = 10.98EE291 pKa = 4.4NVPPAVQEE299 pKa = 4.26AVATTSKK306 pKa = 10.54QPTRR310 pKa = 11.84AEE312 pKa = 4.05EE313 pKa = 4.26EE314 pKa = 4.15EE315 pKa = 4.27LQDD318 pKa = 3.67FQVVKK323 pKa = 10.66KK324 pKa = 10.18KK325 pKa = 9.94PRR327 pKa = 11.84KK328 pKa = 9.45AKK330 pKa = 10.56SSS332 pKa = 3.41
Molecular weight: 37.36 kDa
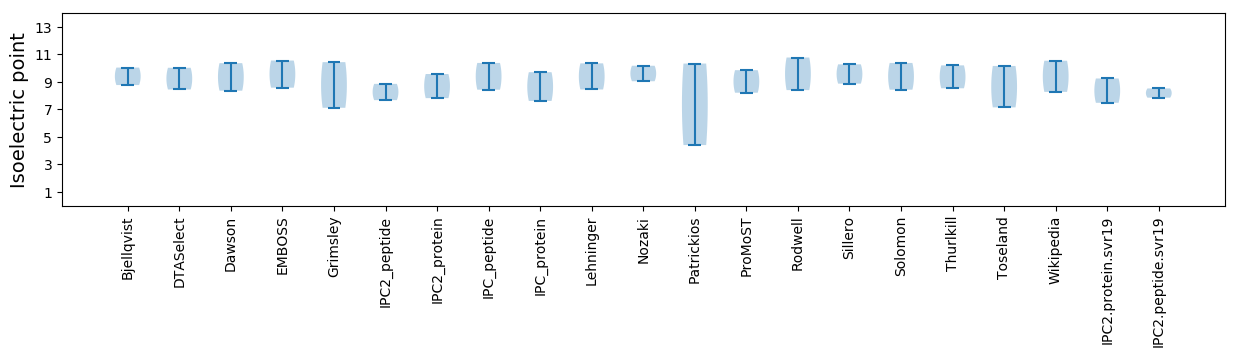
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0A2Q4|K0A2Q4_9VIRU Replication-associated protein OS=Dragonfly circularisvirus OX=1234872 PE=4 SV=1
MM1 pKa = 6.75VKK3 pKa = 9.74SWGRR7 pKa = 11.84CSRR10 pKa = 11.84VLQSTTCLIMLSEE23 pKa = 4.39SVLLLLRR30 pKa = 11.84LLVCLICLLKK40 pKa = 10.08MVMRR44 pKa = 11.84AWRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84MYY51 pKa = 10.46RR52 pKa = 11.84LRR54 pKa = 11.84YY55 pKa = 8.25RR56 pKa = 11.84KK57 pKa = 9.26RR58 pKa = 11.84WQQRR62 pKa = 11.84RR63 pKa = 11.84NSRR66 pKa = 11.84LGRR69 pKa = 11.84RR70 pKa = 11.84KK71 pKa = 9.8RR72 pKa = 11.84SYY74 pKa = 9.71KK75 pKa = 8.67TFKK78 pKa = 10.24SSKK81 pKa = 10.16RR82 pKa = 11.84NLAKK86 pKa = 10.63LNQVKK91 pKa = 7.88VTVRR95 pKa = 11.84DD96 pKa = 3.41LDD98 pKa = 3.47YY99 pKa = 11.05HH100 pKa = 6.8IIKK103 pKa = 10.13IPYY106 pKa = 8.05TGGDD110 pKa = 3.31PRR112 pKa = 11.84QIGSEE117 pKa = 3.86NAEE120 pKa = 4.07MFKK123 pKa = 10.5TDD125 pKa = 3.6VPVHH129 pKa = 5.42ALKK132 pKa = 10.38MKK134 pKa = 10.78LFTLNLQRR142 pKa = 11.84YY143 pKa = 8.0KK144 pKa = 10.21WIKK147 pKa = 9.08FNYY150 pKa = 8.06FSFYY154 pKa = 9.21ITADD158 pKa = 3.88CISYY162 pKa = 10.41DD163 pKa = 2.99IYY165 pKa = 10.63PYY167 pKa = 10.7KK168 pKa = 10.55DD169 pKa = 3.56EE170 pKa = 4.03QSKK173 pKa = 9.18TSGLLSPGINEE184 pKa = 4.41FFKK187 pKa = 11.0HH188 pKa = 6.01LPFYY192 pKa = 9.98LQWDD196 pKa = 4.02LDD198 pKa = 3.92VTFDD202 pKa = 4.38NRR204 pKa = 11.84DD205 pKa = 3.49LTEE208 pKa = 3.79VNYY211 pKa = 10.58PNDD214 pKa = 3.12VHH216 pKa = 6.77AKK218 pKa = 9.78KK219 pKa = 10.46IFIGSRR225 pKa = 11.84KK226 pKa = 8.4PAVFKK231 pKa = 10.64YY232 pKa = 9.51VIPQHH237 pKa = 5.27LRR239 pKa = 11.84KK240 pKa = 9.68YY241 pKa = 9.62LPCGVVKK248 pKa = 10.72SQVTTKK254 pKa = 10.43PLPLYY259 pKa = 10.05LQEE262 pKa = 5.11LFKK265 pKa = 11.04SDD267 pKa = 3.49NFRR270 pKa = 11.84VPSVFYY276 pKa = 11.06GGVTNLMRR284 pKa = 11.84ALDD287 pKa = 4.02LFYY290 pKa = 11.04NKK292 pKa = 9.59PSEE295 pKa = 4.23GRR297 pKa = 11.84DD298 pKa = 3.32VPLRR302 pKa = 11.84FVLTVKK308 pKa = 10.51CYY310 pKa = 11.36ANVTFKK316 pKa = 11.33GIDD319 pKa = 3.4LVV321 pKa = 3.86
MM1 pKa = 6.75VKK3 pKa = 9.74SWGRR7 pKa = 11.84CSRR10 pKa = 11.84VLQSTTCLIMLSEE23 pKa = 4.39SVLLLLRR30 pKa = 11.84LLVCLICLLKK40 pKa = 10.08MVMRR44 pKa = 11.84AWRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84MYY51 pKa = 10.46RR52 pKa = 11.84LRR54 pKa = 11.84YY55 pKa = 8.25RR56 pKa = 11.84KK57 pKa = 9.26RR58 pKa = 11.84WQQRR62 pKa = 11.84RR63 pKa = 11.84NSRR66 pKa = 11.84LGRR69 pKa = 11.84RR70 pKa = 11.84KK71 pKa = 9.8RR72 pKa = 11.84SYY74 pKa = 9.71KK75 pKa = 8.67TFKK78 pKa = 10.24SSKK81 pKa = 10.16RR82 pKa = 11.84NLAKK86 pKa = 10.63LNQVKK91 pKa = 7.88VTVRR95 pKa = 11.84DD96 pKa = 3.41LDD98 pKa = 3.47YY99 pKa = 11.05HH100 pKa = 6.8IIKK103 pKa = 10.13IPYY106 pKa = 8.05TGGDD110 pKa = 3.31PRR112 pKa = 11.84QIGSEE117 pKa = 3.86NAEE120 pKa = 4.07MFKK123 pKa = 10.5TDD125 pKa = 3.6VPVHH129 pKa = 5.42ALKK132 pKa = 10.38MKK134 pKa = 10.78LFTLNLQRR142 pKa = 11.84YY143 pKa = 8.0KK144 pKa = 10.21WIKK147 pKa = 9.08FNYY150 pKa = 8.06FSFYY154 pKa = 9.21ITADD158 pKa = 3.88CISYY162 pKa = 10.41DD163 pKa = 2.99IYY165 pKa = 10.63PYY167 pKa = 10.7KK168 pKa = 10.55DD169 pKa = 3.56EE170 pKa = 4.03QSKK173 pKa = 9.18TSGLLSPGINEE184 pKa = 4.41FFKK187 pKa = 11.0HH188 pKa = 6.01LPFYY192 pKa = 9.98LQWDD196 pKa = 4.02LDD198 pKa = 3.92VTFDD202 pKa = 4.38NRR204 pKa = 11.84DD205 pKa = 3.49LTEE208 pKa = 3.79VNYY211 pKa = 10.58PNDD214 pKa = 3.12VHH216 pKa = 6.77AKK218 pKa = 9.78KK219 pKa = 10.46IFIGSRR225 pKa = 11.84KK226 pKa = 8.4PAVFKK231 pKa = 10.64YY232 pKa = 9.51VIPQHH237 pKa = 5.27LRR239 pKa = 11.84KK240 pKa = 9.68YY241 pKa = 9.62LPCGVVKK248 pKa = 10.72SQVTTKK254 pKa = 10.43PLPLYY259 pKa = 10.05LQEE262 pKa = 5.11LFKK265 pKa = 11.04SDD267 pKa = 3.49NFRR270 pKa = 11.84VPSVFYY276 pKa = 11.06GGVTNLMRR284 pKa = 11.84ALDD287 pKa = 4.02LFYY290 pKa = 11.04NKK292 pKa = 9.59PSEE295 pKa = 4.23GRR297 pKa = 11.84DD298 pKa = 3.32VPLRR302 pKa = 11.84FVLTVKK308 pKa = 10.51CYY310 pKa = 11.36ANVTFKK316 pKa = 11.33GIDD319 pKa = 3.4LVV321 pKa = 3.86
Molecular weight: 38.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
653 |
321 |
332 |
326.5 |
37.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.441 ± 1.245 | 3.063 ± 0.671 |
5.666 ± 0.518 | 4.288 ± 1.366 |
5.36 ± 0.188 | 5.36 ± 0.996 |
1.378 ± 0.136 | 4.9 ± 0.173 |
9.035 ± 0.001 | 9.648 ± 1.902 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.614 | 3.828 ± 0.405 |
5.207 ± 0.406 | 3.063 ± 0.277 |
7.198 ± 1.16 | 7.044 ± 0.619 |
5.207 ± 0.169 | 7.351 ± 0.57 |
1.685 ± 0.097 | 4.594 ± 1.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
