
Accumulibacter phosphatis (strain UW-1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Accumulibacter; Candidatus Accumulibacter phosphatis
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
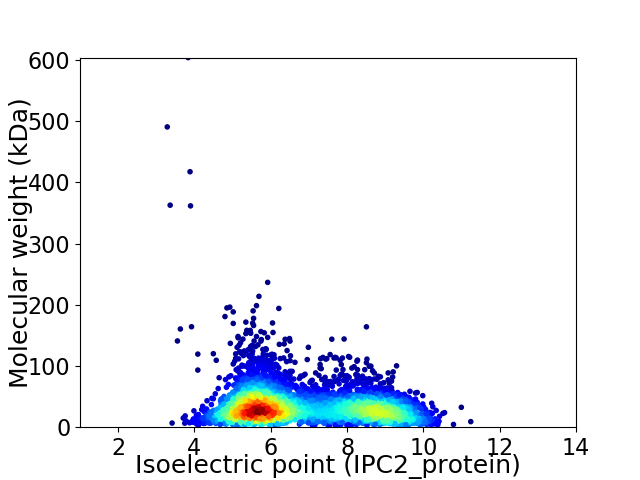
Virtual 2D-PAGE plot for 4438 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7RNV6|C7RNV6_ACCPU Acetyl-CoA acetyltransferase OS=Accumulibacter phosphatis (strain UW-1) OX=522306 GN=CAP2UW1_2144 PE=3 SV=1
MM1 pKa = 8.02LIRR4 pKa = 11.84ITYY7 pKa = 8.54AFPTAASGIYY17 pKa = 10.04GSQEE21 pKa = 3.32LSGFVGLNAQQQAQAEE37 pKa = 4.41RR38 pKa = 11.84ALQTWDD44 pKa = 4.52DD45 pKa = 4.96LIAPDD50 pKa = 4.16MVRR53 pKa = 11.84TTATTSNIEE62 pKa = 4.29FGTSTTGVSYY72 pKa = 11.03AHH74 pKa = 7.38AYY76 pKa = 9.83FPTTGSVWFSRR87 pKa = 11.84SYY89 pKa = 11.41ADD91 pKa = 4.73LMSPQVGQHH100 pKa = 5.33SFMAYY105 pKa = 6.97VHH107 pKa = 6.84EE108 pKa = 5.13IGHH111 pKa = 6.6ALGLDD116 pKa = 3.23HH117 pKa = 6.94MGDD120 pKa = 4.13YY121 pKa = 10.66NGSGSWTPSSYY132 pKa = 10.82EE133 pKa = 3.66DD134 pKa = 3.22SGVYY138 pKa = 10.3SVMSYY143 pKa = 10.07FGPNMNSGSGQVAWADD159 pKa = 3.29WVGADD164 pKa = 3.8GKK166 pKa = 10.57LYY168 pKa = 10.7SPQTPMVNDD177 pKa = 3.11VMAIQAIYY185 pKa = 9.85GAEE188 pKa = 4.09TTTRR192 pKa = 11.84TGDD195 pKa = 3.24TVYY198 pKa = 10.97GFGSNVTGAMAPIFDD213 pKa = 4.2FTLNANPIVTIYY225 pKa = 10.91DD226 pKa = 3.4SAGIDD231 pKa = 3.6TLNLSGWNTACTINLAPGSYY251 pKa = 10.44SSGNSMTYY259 pKa = 10.5NIAIAYY265 pKa = 6.83TCDD268 pKa = 2.95IEE270 pKa = 4.4NAVGGSAGDD279 pKa = 3.62QITGNLLANRR289 pKa = 11.84LDD291 pKa = 3.91GGAGNDD297 pKa = 3.75TLSGGGGDD305 pKa = 4.04DD306 pKa = 3.62VLVAGAGDD314 pKa = 3.82DD315 pKa = 3.76VLDD318 pKa = 4.51GGEE321 pKa = 4.33GTDD324 pKa = 3.24SVVFSGTWGAYY335 pKa = 10.01SYY337 pKa = 10.87TCTNLYY343 pKa = 8.75ATTFAFTFYY352 pKa = 10.91GAATGTDD359 pKa = 3.43TLRR362 pKa = 11.84GIEE365 pKa = 4.22TFVFADD371 pKa = 3.39VTKK374 pKa = 10.62YY375 pKa = 10.88ASEE378 pKa = 4.08LSGAGPSPSLVSISSSAATQAEE400 pKa = 4.7GNAGQTLFSYY410 pKa = 10.31KK411 pKa = 10.21VSLSAASSSVQTVNWSLSGTGSSAADD437 pKa = 3.54PASDD441 pKa = 4.24FSGATSGSITFQVGEE456 pKa = 4.33TEE458 pKa = 4.06KK459 pKa = 10.4TIQIYY464 pKa = 10.78ARR466 pKa = 11.84GDD468 pKa = 3.46TSVEE472 pKa = 3.8ADD474 pKa = 3.16EE475 pKa = 5.44HH476 pKa = 5.57FTVTLSNPSAGLKK489 pKa = 9.56FASSSAQGTILNDD502 pKa = 3.64DD503 pKa = 3.96TSSGGDD509 pKa = 3.38DD510 pKa = 3.92FGSTTASAGNLAVNGGAVSGAIEE533 pKa = 4.0RR534 pKa = 11.84AGDD537 pKa = 3.52LDD539 pKa = 4.14LFRR542 pKa = 11.84VNLNAGVTYY551 pKa = 10.89VFDD554 pKa = 4.52LVATGSGVDD563 pKa = 4.19PYY565 pKa = 11.43LSLYY569 pKa = 8.98ATTATSLSAVASNDD583 pKa = 2.84NVRR586 pKa = 11.84AGTSSAQITYY596 pKa = 7.72TASSSATYY604 pKa = 10.77YY605 pKa = 11.28LLARR609 pKa = 11.84DD610 pKa = 3.67TWTGTGSYY618 pKa = 9.02TISAITFAGRR628 pKa = 11.84TITGDD633 pKa = 3.03EE634 pKa = 3.92AANRR638 pKa = 11.84LVGTLGDD645 pKa = 3.75DD646 pKa = 3.44VLYY649 pKa = 10.99GLGGADD655 pKa = 3.55VLSGDD660 pKa = 4.31GGDD663 pKa = 4.33DD664 pKa = 3.56RR665 pKa = 11.84LDD667 pKa = 3.89GGSGADD673 pKa = 3.84AMSGGNGNDD682 pKa = 2.79IYY684 pKa = 11.68VVDD687 pKa = 4.26NVADD691 pKa = 4.14SVSEE695 pKa = 3.93TSAAGGIDD703 pKa = 3.06RR704 pKa = 11.84VEE706 pKa = 4.3SSVSFTLGTNLEE718 pKa = 4.15NLVLTGNADD727 pKa = 3.59SNGTGNSQNNQLTGNSGNNILDD749 pKa = 3.85GKK751 pKa = 10.83AGLDD755 pKa = 3.54TFAGGLGNDD764 pKa = 3.63TYY766 pKa = 11.5LLDD769 pKa = 4.5MEE771 pKa = 5.6GEE773 pKa = 4.2LANVTEE779 pKa = 4.45NAGQGTDD786 pKa = 3.21TLTVVYY792 pKa = 10.37ANTSAVKK799 pKa = 10.5SIALTGNLANIEE811 pKa = 4.07NVLLSGAGLFNVLGNGLANALTGNASVNRR840 pKa = 11.84LEE842 pKa = 4.51GAAGNDD848 pKa = 3.57TLDD851 pKa = 3.9GQGGADD857 pKa = 3.62QLIGGQGDD865 pKa = 3.76DD866 pKa = 3.78VYY868 pKa = 11.48KK869 pKa = 10.57VDD871 pKa = 3.83QLGDD875 pKa = 3.71VLIEE879 pKa = 4.29NADD882 pKa = 3.77EE883 pKa = 4.92GSDD886 pKa = 3.61TVLVALSASGTSYY899 pKa = 11.57VLGNHH904 pKa = 6.85LEE906 pKa = 4.21NATVVAAVAINLTGNDD922 pKa = 3.84LANVLTGNALANRR935 pKa = 11.84LDD937 pKa = 3.84GGAGIDD943 pKa = 3.81TLIGGAGGDD952 pKa = 3.85TYY954 pKa = 11.71VVDD957 pKa = 3.5QVGDD961 pKa = 3.78VLVEE965 pKa = 4.11TSSLASEE972 pKa = 4.28IDD974 pKa = 3.43RR975 pKa = 11.84VEE977 pKa = 4.9SYY979 pKa = 11.58VDD981 pKa = 3.73WILGANLEE989 pKa = 4.03NLTLLGNGNLAGTGNALNNTLIGNAGANRR1018 pKa = 11.84LDD1020 pKa = 3.92GGQGADD1026 pKa = 3.49ILLGGAGDD1034 pKa = 3.91DD1035 pKa = 3.91RR1036 pKa = 11.84YY1037 pKa = 11.31VVDD1040 pKa = 3.81NLGDD1044 pKa = 3.59KK1045 pKa = 10.73VYY1047 pKa = 8.86EE1048 pKa = 4.45TTTSTSAYY1056 pKa = 9.56DD1057 pKa = 3.23AGGIDD1062 pKa = 4.11TVDD1065 pKa = 3.83SSINWTLGNFVEE1077 pKa = 4.63NLTLTGSGNLSGIGNALANTLRR1099 pKa = 11.84GNAGINVLDD1108 pKa = 4.5GKK1110 pKa = 10.89AGVDD1114 pKa = 3.49VLDD1117 pKa = 4.49GGEE1120 pKa = 4.15GSDD1123 pKa = 3.3IYY1125 pKa = 11.39LVGLASDD1132 pKa = 4.0HH1133 pKa = 6.4AAAEE1137 pKa = 4.38IADD1140 pKa = 4.02SGNGGSDD1147 pKa = 3.22EE1148 pKa = 4.16VRR1150 pKa = 11.84FAASSSGTLTLFAGDD1165 pKa = 3.53TGIEE1169 pKa = 3.89RR1170 pKa = 11.84VVIGSGLGEE1179 pKa = 4.1VAVSSGTLALDD1190 pKa = 3.43VDD1192 pKa = 4.49ASRR1195 pKa = 11.84VANALSVLGNAGANVLTGTAYY1216 pKa = 10.48ADD1218 pKa = 4.15RR1219 pKa = 11.84LDD1221 pKa = 4.24GGAGADD1227 pKa = 3.65RR1228 pKa = 11.84LIGGGGADD1236 pKa = 3.82TLIGGAGNDD1245 pKa = 3.56VYY1247 pKa = 11.66VVDD1250 pKa = 4.7SGDD1253 pKa = 4.12AIIEE1257 pKa = 4.23ASTLASEE1264 pKa = 4.49IDD1266 pKa = 3.54RR1267 pKa = 11.84VEE1269 pKa = 4.9SYY1271 pKa = 11.58VDD1273 pKa = 3.73WILGANLEE1281 pKa = 4.08NLTLLGTGNLAGTGNALNNTLTGNAGANRR1310 pKa = 11.84LDD1312 pKa = 3.92GGQGADD1318 pKa = 3.49ILLGGAGDD1326 pKa = 3.91DD1327 pKa = 3.91RR1328 pKa = 11.84YY1329 pKa = 11.31VVDD1332 pKa = 3.81NLGDD1336 pKa = 3.59KK1337 pKa = 10.73VYY1339 pKa = 8.86EE1340 pKa = 4.45TTTSTSQYY1348 pKa = 10.74DD1349 pKa = 3.26AGGTDD1354 pKa = 3.75TVDD1357 pKa = 4.54SSINWTLGNFVEE1369 pKa = 4.63NLTLTGSGNLSGIGNALANTLRR1391 pKa = 11.84GNAGINVLDD1400 pKa = 4.5GKK1402 pKa = 10.89AGVDD1406 pKa = 3.49VLDD1409 pKa = 4.49GGEE1412 pKa = 4.15GSDD1415 pKa = 3.3IYY1417 pKa = 11.39LVGLASDD1424 pKa = 4.0HH1425 pKa = 6.4AAAEE1429 pKa = 4.38IADD1432 pKa = 4.02SGNGGSDD1439 pKa = 3.24EE1440 pKa = 4.24VRR1442 pKa = 11.84FAANTSGTLTLFAGDD1457 pKa = 3.53TGIEE1461 pKa = 3.89RR1462 pKa = 11.84VVIGSGLGEE1471 pKa = 4.1VAVSSGTLALDD1482 pKa = 3.43VDD1484 pKa = 4.49ASRR1487 pKa = 11.84VANALSLLGNAGANVLTGTAYY1508 pKa = 10.65ADD1510 pKa = 3.91TLDD1513 pKa = 4.29GGAGADD1519 pKa = 3.8RR1520 pKa = 11.84LIGGDD1525 pKa = 3.58GNDD1528 pKa = 3.74RR1529 pKa = 11.84LIGGAGNDD1537 pKa = 3.92TLTGGAGADD1546 pKa = 3.34QFVLTQALSSTTNRR1560 pKa = 11.84DD1561 pKa = 2.87VLADD1565 pKa = 4.35FVSGTDD1571 pKa = 3.73TIQLSVSVFRR1581 pKa = 11.84AVTSRR1586 pKa = 11.84DD1587 pKa = 2.38IWNSSTRR1594 pKa = 11.84AIRR1597 pKa = 11.84GG1598 pKa = 3.36
MM1 pKa = 8.02LIRR4 pKa = 11.84ITYY7 pKa = 8.54AFPTAASGIYY17 pKa = 10.04GSQEE21 pKa = 3.32LSGFVGLNAQQQAQAEE37 pKa = 4.41RR38 pKa = 11.84ALQTWDD44 pKa = 4.52DD45 pKa = 4.96LIAPDD50 pKa = 4.16MVRR53 pKa = 11.84TTATTSNIEE62 pKa = 4.29FGTSTTGVSYY72 pKa = 11.03AHH74 pKa = 7.38AYY76 pKa = 9.83FPTTGSVWFSRR87 pKa = 11.84SYY89 pKa = 11.41ADD91 pKa = 4.73LMSPQVGQHH100 pKa = 5.33SFMAYY105 pKa = 6.97VHH107 pKa = 6.84EE108 pKa = 5.13IGHH111 pKa = 6.6ALGLDD116 pKa = 3.23HH117 pKa = 6.94MGDD120 pKa = 4.13YY121 pKa = 10.66NGSGSWTPSSYY132 pKa = 10.82EE133 pKa = 3.66DD134 pKa = 3.22SGVYY138 pKa = 10.3SVMSYY143 pKa = 10.07FGPNMNSGSGQVAWADD159 pKa = 3.29WVGADD164 pKa = 3.8GKK166 pKa = 10.57LYY168 pKa = 10.7SPQTPMVNDD177 pKa = 3.11VMAIQAIYY185 pKa = 9.85GAEE188 pKa = 4.09TTTRR192 pKa = 11.84TGDD195 pKa = 3.24TVYY198 pKa = 10.97GFGSNVTGAMAPIFDD213 pKa = 4.2FTLNANPIVTIYY225 pKa = 10.91DD226 pKa = 3.4SAGIDD231 pKa = 3.6TLNLSGWNTACTINLAPGSYY251 pKa = 10.44SSGNSMTYY259 pKa = 10.5NIAIAYY265 pKa = 6.83TCDD268 pKa = 2.95IEE270 pKa = 4.4NAVGGSAGDD279 pKa = 3.62QITGNLLANRR289 pKa = 11.84LDD291 pKa = 3.91GGAGNDD297 pKa = 3.75TLSGGGGDD305 pKa = 4.04DD306 pKa = 3.62VLVAGAGDD314 pKa = 3.82DD315 pKa = 3.76VLDD318 pKa = 4.51GGEE321 pKa = 4.33GTDD324 pKa = 3.24SVVFSGTWGAYY335 pKa = 10.01SYY337 pKa = 10.87TCTNLYY343 pKa = 8.75ATTFAFTFYY352 pKa = 10.91GAATGTDD359 pKa = 3.43TLRR362 pKa = 11.84GIEE365 pKa = 4.22TFVFADD371 pKa = 3.39VTKK374 pKa = 10.62YY375 pKa = 10.88ASEE378 pKa = 4.08LSGAGPSPSLVSISSSAATQAEE400 pKa = 4.7GNAGQTLFSYY410 pKa = 10.31KK411 pKa = 10.21VSLSAASSSVQTVNWSLSGTGSSAADD437 pKa = 3.54PASDD441 pKa = 4.24FSGATSGSITFQVGEE456 pKa = 4.33TEE458 pKa = 4.06KK459 pKa = 10.4TIQIYY464 pKa = 10.78ARR466 pKa = 11.84GDD468 pKa = 3.46TSVEE472 pKa = 3.8ADD474 pKa = 3.16EE475 pKa = 5.44HH476 pKa = 5.57FTVTLSNPSAGLKK489 pKa = 9.56FASSSAQGTILNDD502 pKa = 3.64DD503 pKa = 3.96TSSGGDD509 pKa = 3.38DD510 pKa = 3.92FGSTTASAGNLAVNGGAVSGAIEE533 pKa = 4.0RR534 pKa = 11.84AGDD537 pKa = 3.52LDD539 pKa = 4.14LFRR542 pKa = 11.84VNLNAGVTYY551 pKa = 10.89VFDD554 pKa = 4.52LVATGSGVDD563 pKa = 4.19PYY565 pKa = 11.43LSLYY569 pKa = 8.98ATTATSLSAVASNDD583 pKa = 2.84NVRR586 pKa = 11.84AGTSSAQITYY596 pKa = 7.72TASSSATYY604 pKa = 10.77YY605 pKa = 11.28LLARR609 pKa = 11.84DD610 pKa = 3.67TWTGTGSYY618 pKa = 9.02TISAITFAGRR628 pKa = 11.84TITGDD633 pKa = 3.03EE634 pKa = 3.92AANRR638 pKa = 11.84LVGTLGDD645 pKa = 3.75DD646 pKa = 3.44VLYY649 pKa = 10.99GLGGADD655 pKa = 3.55VLSGDD660 pKa = 4.31GGDD663 pKa = 4.33DD664 pKa = 3.56RR665 pKa = 11.84LDD667 pKa = 3.89GGSGADD673 pKa = 3.84AMSGGNGNDD682 pKa = 2.79IYY684 pKa = 11.68VVDD687 pKa = 4.26NVADD691 pKa = 4.14SVSEE695 pKa = 3.93TSAAGGIDD703 pKa = 3.06RR704 pKa = 11.84VEE706 pKa = 4.3SSVSFTLGTNLEE718 pKa = 4.15NLVLTGNADD727 pKa = 3.59SNGTGNSQNNQLTGNSGNNILDD749 pKa = 3.85GKK751 pKa = 10.83AGLDD755 pKa = 3.54TFAGGLGNDD764 pKa = 3.63TYY766 pKa = 11.5LLDD769 pKa = 4.5MEE771 pKa = 5.6GEE773 pKa = 4.2LANVTEE779 pKa = 4.45NAGQGTDD786 pKa = 3.21TLTVVYY792 pKa = 10.37ANTSAVKK799 pKa = 10.5SIALTGNLANIEE811 pKa = 4.07NVLLSGAGLFNVLGNGLANALTGNASVNRR840 pKa = 11.84LEE842 pKa = 4.51GAAGNDD848 pKa = 3.57TLDD851 pKa = 3.9GQGGADD857 pKa = 3.62QLIGGQGDD865 pKa = 3.76DD866 pKa = 3.78VYY868 pKa = 11.48KK869 pKa = 10.57VDD871 pKa = 3.83QLGDD875 pKa = 3.71VLIEE879 pKa = 4.29NADD882 pKa = 3.77EE883 pKa = 4.92GSDD886 pKa = 3.61TVLVALSASGTSYY899 pKa = 11.57VLGNHH904 pKa = 6.85LEE906 pKa = 4.21NATVVAAVAINLTGNDD922 pKa = 3.84LANVLTGNALANRR935 pKa = 11.84LDD937 pKa = 3.84GGAGIDD943 pKa = 3.81TLIGGAGGDD952 pKa = 3.85TYY954 pKa = 11.71VVDD957 pKa = 3.5QVGDD961 pKa = 3.78VLVEE965 pKa = 4.11TSSLASEE972 pKa = 4.28IDD974 pKa = 3.43RR975 pKa = 11.84VEE977 pKa = 4.9SYY979 pKa = 11.58VDD981 pKa = 3.73WILGANLEE989 pKa = 4.03NLTLLGNGNLAGTGNALNNTLIGNAGANRR1018 pKa = 11.84LDD1020 pKa = 3.92GGQGADD1026 pKa = 3.49ILLGGAGDD1034 pKa = 3.91DD1035 pKa = 3.91RR1036 pKa = 11.84YY1037 pKa = 11.31VVDD1040 pKa = 3.81NLGDD1044 pKa = 3.59KK1045 pKa = 10.73VYY1047 pKa = 8.86EE1048 pKa = 4.45TTTSTSAYY1056 pKa = 9.56DD1057 pKa = 3.23AGGIDD1062 pKa = 4.11TVDD1065 pKa = 3.83SSINWTLGNFVEE1077 pKa = 4.63NLTLTGSGNLSGIGNALANTLRR1099 pKa = 11.84GNAGINVLDD1108 pKa = 4.5GKK1110 pKa = 10.89AGVDD1114 pKa = 3.49VLDD1117 pKa = 4.49GGEE1120 pKa = 4.15GSDD1123 pKa = 3.3IYY1125 pKa = 11.39LVGLASDD1132 pKa = 4.0HH1133 pKa = 6.4AAAEE1137 pKa = 4.38IADD1140 pKa = 4.02SGNGGSDD1147 pKa = 3.22EE1148 pKa = 4.16VRR1150 pKa = 11.84FAASSSGTLTLFAGDD1165 pKa = 3.53TGIEE1169 pKa = 3.89RR1170 pKa = 11.84VVIGSGLGEE1179 pKa = 4.1VAVSSGTLALDD1190 pKa = 3.43VDD1192 pKa = 4.49ASRR1195 pKa = 11.84VANALSVLGNAGANVLTGTAYY1216 pKa = 10.48ADD1218 pKa = 4.15RR1219 pKa = 11.84LDD1221 pKa = 4.24GGAGADD1227 pKa = 3.65RR1228 pKa = 11.84LIGGGGADD1236 pKa = 3.82TLIGGAGNDD1245 pKa = 3.56VYY1247 pKa = 11.66VVDD1250 pKa = 4.7SGDD1253 pKa = 4.12AIIEE1257 pKa = 4.23ASTLASEE1264 pKa = 4.49IDD1266 pKa = 3.54RR1267 pKa = 11.84VEE1269 pKa = 4.9SYY1271 pKa = 11.58VDD1273 pKa = 3.73WILGANLEE1281 pKa = 4.08NLTLLGTGNLAGTGNALNNTLTGNAGANRR1310 pKa = 11.84LDD1312 pKa = 3.92GGQGADD1318 pKa = 3.49ILLGGAGDD1326 pKa = 3.91DD1327 pKa = 3.91RR1328 pKa = 11.84YY1329 pKa = 11.31VVDD1332 pKa = 3.81NLGDD1336 pKa = 3.59KK1337 pKa = 10.73VYY1339 pKa = 8.86EE1340 pKa = 4.45TTTSTSQYY1348 pKa = 10.74DD1349 pKa = 3.26AGGTDD1354 pKa = 3.75TVDD1357 pKa = 4.54SSINWTLGNFVEE1369 pKa = 4.63NLTLTGSGNLSGIGNALANTLRR1391 pKa = 11.84GNAGINVLDD1400 pKa = 4.5GKK1402 pKa = 10.89AGVDD1406 pKa = 3.49VLDD1409 pKa = 4.49GGEE1412 pKa = 4.15GSDD1415 pKa = 3.3IYY1417 pKa = 11.39LVGLASDD1424 pKa = 4.0HH1425 pKa = 6.4AAAEE1429 pKa = 4.38IADD1432 pKa = 4.02SGNGGSDD1439 pKa = 3.24EE1440 pKa = 4.24VRR1442 pKa = 11.84FAANTSGTLTLFAGDD1457 pKa = 3.53TGIEE1461 pKa = 3.89RR1462 pKa = 11.84VVIGSGLGEE1471 pKa = 4.1VAVSSGTLALDD1482 pKa = 3.43VDD1484 pKa = 4.49ASRR1487 pKa = 11.84VANALSLLGNAGANVLTGTAYY1508 pKa = 10.65ADD1510 pKa = 3.91TLDD1513 pKa = 4.29GGAGADD1519 pKa = 3.8RR1520 pKa = 11.84LIGGDD1525 pKa = 3.58GNDD1528 pKa = 3.74RR1529 pKa = 11.84LIGGAGNDD1537 pKa = 3.92TLTGGAGADD1546 pKa = 3.34QFVLTQALSSTTNRR1560 pKa = 11.84DD1561 pKa = 2.87VLADD1565 pKa = 4.35FVSGTDD1571 pKa = 3.73TIQLSVSVFRR1581 pKa = 11.84AVTSRR1586 pKa = 11.84DD1587 pKa = 2.38IWNSSTRR1594 pKa = 11.84AIRR1597 pKa = 11.84GG1598 pKa = 3.36
Molecular weight: 160.52 kDa
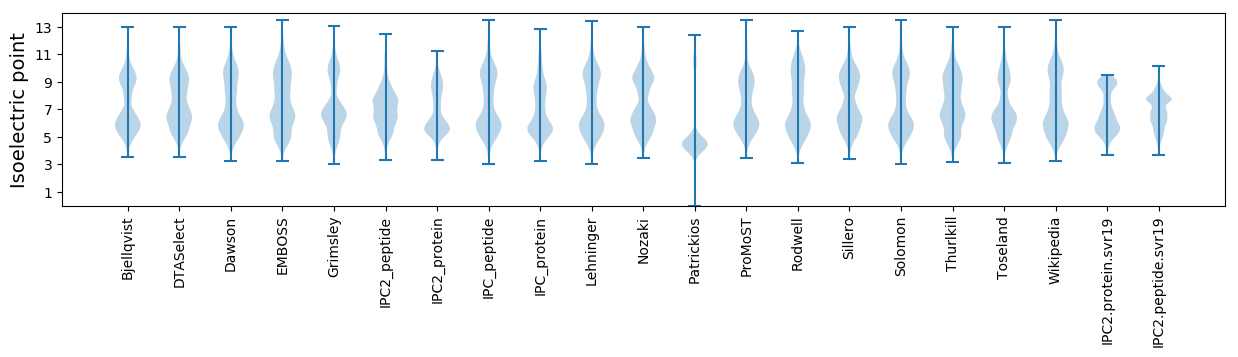
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7RSN1|C7RSN1_ACCPU von Willebrand factor type A OS=Accumulibacter phosphatis (strain UW-1) OX=522306 GN=CAP2UW1_2647 PE=4 SV=1
MM1 pKa = 7.71ARR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 10.06KK6 pKa = 10.25PSRR9 pKa = 11.84LLLLQLLLPLPLLPLLLRR27 pKa = 11.84LPLLPLLLRR36 pKa = 11.84LLRR39 pKa = 11.84PRR41 pKa = 11.84MLRR44 pKa = 11.84SPPMPPNPPTPPWTRR59 pKa = 11.84PRR61 pKa = 11.84KK62 pKa = 9.9LPALLRR68 pKa = 11.84KK69 pKa = 9.28ALPMQPKK76 pKa = 8.11MQQ78 pKa = 4.02
MM1 pKa = 7.71ARR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 10.06KK6 pKa = 10.25PSRR9 pKa = 11.84LLLLQLLLPLPLLPLLLRR27 pKa = 11.84LPLLPLLLRR36 pKa = 11.84LLRR39 pKa = 11.84PRR41 pKa = 11.84MLRR44 pKa = 11.84SPPMPPNPPTPPWTRR59 pKa = 11.84PRR61 pKa = 11.84KK62 pKa = 9.9LPALLRR68 pKa = 11.84KK69 pKa = 9.28ALPMQPKK76 pKa = 8.11MQQ78 pKa = 4.02
Molecular weight: 9.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1504008 |
30 |
5854 |
338.9 |
36.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.498 ± 0.046 | 1.023 ± 0.013 |
5.49 ± 0.05 | 5.538 ± 0.04 |
3.614 ± 0.023 | 8.18 ± 0.065 |
2.173 ± 0.02 | 4.592 ± 0.023 |
2.98 ± 0.032 | 11.359 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.131 ± 0.023 | 2.538 ± 0.027 |
5.038 ± 0.032 | 3.647 ± 0.028 |
7.551 ± 0.046 | 5.542 ± 0.029 |
4.951 ± 0.048 | 7.461 ± 0.039 |
1.426 ± 0.016 | 2.269 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
