
Stemphylium lycopersici
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Pleosporineae; Pleosporaceae; Stemphylium
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 9079 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
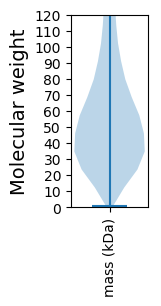
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A364N1R0|A0A364N1R0_9PLEO 3'(2') 5'-bisphosphate nucleotidase OS=Stemphylium lycopersici OX=183478 GN=DDE83_005700 PE=3 SV=1
MM1 pKa = 7.56GFLSEE6 pKa = 5.51LISDD10 pKa = 4.66FKK12 pKa = 11.33GSSSSGQGDD21 pKa = 3.45QQQYY25 pKa = 7.46QQQQSGYY32 pKa = 8.11SQYY35 pKa = 10.82PPQQHH40 pKa = 6.18SPQPPPVSPPWYY52 pKa = 10.12AEE54 pKa = 3.24WDD56 pKa = 3.64SRR58 pKa = 11.84DD59 pKa = 3.57NRR61 pKa = 11.84WFFVNQHH68 pKa = 4.82TGEE71 pKa = 3.89RR72 pKa = 11.84TYY74 pKa = 11.12NYY76 pKa = 8.05PGPGYY81 pKa = 10.39AQQQGDD87 pKa = 4.14YY88 pKa = 10.36GAPPPQGGYY97 pKa = 10.98GYY99 pKa = 10.91GQGGYY104 pKa = 9.44SNQGAYY110 pKa = 8.34EE111 pKa = 3.94QSAYY115 pKa = 10.9DD116 pKa = 4.09PTRR119 pKa = 11.84SAQPQEE125 pKa = 4.31EE126 pKa = 4.35KK127 pKa = 10.89SGGNGWKK134 pKa = 10.01YY135 pKa = 10.66AAAGAAGLAGGALLMHH151 pKa = 7.09EE152 pKa = 4.55SDD154 pKa = 4.96KK155 pKa = 11.49IEE157 pKa = 5.32DD158 pKa = 3.72GWDD161 pKa = 3.28SAKK164 pKa = 10.81QNISNGVDD172 pKa = 3.01EE173 pKa = 5.35FGNDD177 pKa = 2.95VSNFPEE183 pKa = 4.46NAAGWTGEE191 pKa = 3.98QVGRR195 pKa = 11.84AEE197 pKa = 4.13NFGDD201 pKa = 4.81DD202 pKa = 5.56VEE204 pKa = 4.72QKK206 pKa = 10.13WDD208 pKa = 3.9DD209 pKa = 3.58GVQDD213 pKa = 4.27VEE215 pKa = 5.22DD216 pKa = 4.61FPEE219 pKa = 4.75NAAEE223 pKa = 4.01WTGEE227 pKa = 4.0KK228 pKa = 10.02VQEE231 pKa = 4.45VEE233 pKa = 6.29DD234 pKa = 3.98IPQDD238 pKa = 4.93IEE240 pKa = 4.54NKK242 pKa = 7.83WDD244 pKa = 3.4NMTEE248 pKa = 4.27GIEE251 pKa = 4.44QFGDD255 pKa = 3.8DD256 pKa = 4.41MGDD259 pKa = 3.29AYY261 pKa = 11.01DD262 pKa = 4.0EE263 pKa = 4.69GRR265 pKa = 11.84DD266 pKa = 3.59EE267 pKa = 5.28QRR269 pKa = 11.84YY270 pKa = 8.89EE271 pKa = 4.05DD272 pKa = 5.21DD273 pKa = 3.71YY274 pKa = 12.24
MM1 pKa = 7.56GFLSEE6 pKa = 5.51LISDD10 pKa = 4.66FKK12 pKa = 11.33GSSSSGQGDD21 pKa = 3.45QQQYY25 pKa = 7.46QQQQSGYY32 pKa = 8.11SQYY35 pKa = 10.82PPQQHH40 pKa = 6.18SPQPPPVSPPWYY52 pKa = 10.12AEE54 pKa = 3.24WDD56 pKa = 3.64SRR58 pKa = 11.84DD59 pKa = 3.57NRR61 pKa = 11.84WFFVNQHH68 pKa = 4.82TGEE71 pKa = 3.89RR72 pKa = 11.84TYY74 pKa = 11.12NYY76 pKa = 8.05PGPGYY81 pKa = 10.39AQQQGDD87 pKa = 4.14YY88 pKa = 10.36GAPPPQGGYY97 pKa = 10.98GYY99 pKa = 10.91GQGGYY104 pKa = 9.44SNQGAYY110 pKa = 8.34EE111 pKa = 3.94QSAYY115 pKa = 10.9DD116 pKa = 4.09PTRR119 pKa = 11.84SAQPQEE125 pKa = 4.31EE126 pKa = 4.35KK127 pKa = 10.89SGGNGWKK134 pKa = 10.01YY135 pKa = 10.66AAAGAAGLAGGALLMHH151 pKa = 7.09EE152 pKa = 4.55SDD154 pKa = 4.96KK155 pKa = 11.49IEE157 pKa = 5.32DD158 pKa = 3.72GWDD161 pKa = 3.28SAKK164 pKa = 10.81QNISNGVDD172 pKa = 3.01EE173 pKa = 5.35FGNDD177 pKa = 2.95VSNFPEE183 pKa = 4.46NAAGWTGEE191 pKa = 3.98QVGRR195 pKa = 11.84AEE197 pKa = 4.13NFGDD201 pKa = 4.81DD202 pKa = 5.56VEE204 pKa = 4.72QKK206 pKa = 10.13WDD208 pKa = 3.9DD209 pKa = 3.58GVQDD213 pKa = 4.27VEE215 pKa = 5.22DD216 pKa = 4.61FPEE219 pKa = 4.75NAAEE223 pKa = 4.01WTGEE227 pKa = 4.0KK228 pKa = 10.02VQEE231 pKa = 4.45VEE233 pKa = 6.29DD234 pKa = 3.98IPQDD238 pKa = 4.93IEE240 pKa = 4.54NKK242 pKa = 7.83WDD244 pKa = 3.4NMTEE248 pKa = 4.27GIEE251 pKa = 4.44QFGDD255 pKa = 3.8DD256 pKa = 4.41MGDD259 pKa = 3.29AYY261 pKa = 11.01DD262 pKa = 4.0EE263 pKa = 4.69GRR265 pKa = 11.84DD266 pKa = 3.59EE267 pKa = 5.28QRR269 pKa = 11.84YY270 pKa = 8.89EE271 pKa = 4.05DD272 pKa = 5.21DD273 pKa = 3.71YY274 pKa = 12.24
Molecular weight: 30.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A364N615|A0A364N615_9PLEO Replication factor A protein 3 OS=Stemphylium lycopersici OX=183478 GN=DDE83_003943 PE=3 SV=1
MM1 pKa = 7.54PLARR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.57APRR10 pKa = 11.84TARR13 pKa = 11.84ATRR16 pKa = 11.84TTARR20 pKa = 11.84PSLKK24 pKa = 8.96TRR26 pKa = 11.84LMGGRR31 pKa = 11.84KK32 pKa = 7.3TRR34 pKa = 11.84TKK36 pKa = 9.17RR37 pKa = 11.84TPGTTVTTTTTTRR50 pKa = 11.84TTRR53 pKa = 11.84TAGGHH58 pKa = 6.24HH59 pKa = 6.1SHH61 pKa = 7.22AAPVHH66 pKa = 4.45HH67 pKa = 7.29HH68 pKa = 6.34KK69 pKa = 10.63RR70 pKa = 11.84HH71 pKa = 4.81ATMGDD76 pKa = 3.45KK77 pKa = 10.91VSGMMMKK84 pKa = 10.69LKK86 pKa = 10.71GSLTHH91 pKa = 7.24KK92 pKa = 10.05PGLKK96 pKa = 10.08AAGTRR101 pKa = 11.84RR102 pKa = 11.84MHH104 pKa = 5.92GTDD107 pKa = 2.72GRR109 pKa = 11.84NARR112 pKa = 11.84RR113 pKa = 11.84VYY115 pKa = 10.89
MM1 pKa = 7.54PLARR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 5.57APRR10 pKa = 11.84TARR13 pKa = 11.84ATRR16 pKa = 11.84TTARR20 pKa = 11.84PSLKK24 pKa = 8.96TRR26 pKa = 11.84LMGGRR31 pKa = 11.84KK32 pKa = 7.3TRR34 pKa = 11.84TKK36 pKa = 9.17RR37 pKa = 11.84TPGTTVTTTTTTRR50 pKa = 11.84TTRR53 pKa = 11.84TAGGHH58 pKa = 6.24HH59 pKa = 6.1SHH61 pKa = 7.22AAPVHH66 pKa = 4.45HH67 pKa = 7.29HH68 pKa = 6.34KK69 pKa = 10.63RR70 pKa = 11.84HH71 pKa = 4.81ATMGDD76 pKa = 3.45KK77 pKa = 10.91VSGMMMKK84 pKa = 10.69LKK86 pKa = 10.71GSLTHH91 pKa = 7.24KK92 pKa = 10.05PGLKK96 pKa = 10.08AAGTRR101 pKa = 11.84RR102 pKa = 11.84MHH104 pKa = 5.92GTDD107 pKa = 2.72GRR109 pKa = 11.84NARR112 pKa = 11.84RR113 pKa = 11.84VYY115 pKa = 10.89
Molecular weight: 12.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5419111 |
9 |
10241 |
596.9 |
65.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.957 ± 0.018 | 1.151 ± 0.008 |
5.728 ± 0.017 | 6.271 ± 0.023 |
3.632 ± 0.015 | 6.822 ± 0.02 |
2.448 ± 0.009 | 4.716 ± 0.016 |
5.051 ± 0.021 | 8.454 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.009 | 3.751 ± 0.013 |
6.262 ± 0.03 | 4.179 ± 0.02 |
5.978 ± 0.019 | 8.121 ± 0.028 |
6.073 ± 0.016 | 5.938 ± 0.016 |
1.434 ± 0.008 | 2.778 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
