
Beihai narna-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.29
Get precalculated fractions of proteins
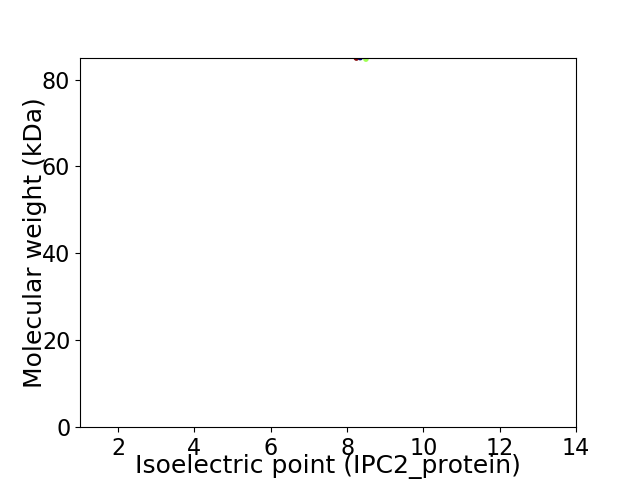
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIB0|A0A1L3KIB0_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 12 OX=1922439 PE=4 SV=1
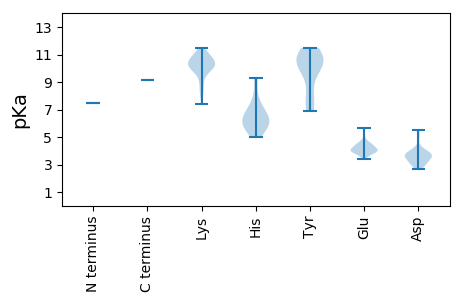
MM1 pKa = 7.48SKK3 pKa = 10.62SDD5 pKa = 3.94DD6 pKa = 3.69RR7 pKa = 11.84RR8 pKa = 11.84PSDD11 pKa = 3.02SQYY14 pKa = 11.06PLRR17 pKa = 11.84LSVSKK22 pKa = 9.82TKK24 pKa = 10.67DD25 pKa = 2.93EE26 pKa = 3.92EE27 pKa = 4.15AFRR30 pKa = 11.84VGLLQEE36 pKa = 3.94PRR38 pKa = 11.84CRR40 pKa = 11.84KK41 pKa = 9.3VALLLAPSCKK51 pKa = 9.74VEE53 pKa = 4.52SVATTSPADD62 pKa = 3.58PLKK65 pKa = 10.96PPGADD70 pKa = 2.7NWRR73 pKa = 11.84LSPSDD78 pKa = 3.55GQIVTTLKK86 pKa = 10.03IVSIITAAIEE96 pKa = 4.18DD97 pKa = 3.68VHH99 pKa = 7.73DD100 pKa = 3.6FHH102 pKa = 9.26LNDD105 pKa = 3.63RR106 pKa = 11.84QIVQAVLMWLPYY118 pKa = 10.9VMADD122 pKa = 2.66VWGPVLKK129 pKa = 10.49ARR131 pKa = 11.84KK132 pKa = 9.58DD133 pKa = 3.3ILFAYY138 pKa = 10.1ALRR141 pKa = 11.84QEE143 pKa = 5.07EE144 pKa = 5.46IPPPWCEE151 pKa = 4.25CCNQTYY157 pKa = 10.41ILPGRR162 pKa = 11.84AGSALRR168 pKa = 11.84QRR170 pKa = 11.84VLYY173 pKa = 10.31NKK175 pKa = 10.33NEE177 pKa = 4.31DD178 pKa = 3.26NRR180 pKa = 11.84IFANTILHH188 pKa = 5.82GLKK191 pKa = 10.27KK192 pKa = 10.58GMPKK196 pKa = 10.31LSEE199 pKa = 3.7AHH201 pKa = 6.85VIDD204 pKa = 5.49GYY206 pKa = 10.87LKK208 pKa = 10.07HH209 pKa = 6.9RR210 pKa = 11.84EE211 pKa = 3.96RR212 pKa = 11.84LSTDD216 pKa = 2.92KK217 pKa = 11.5GPVDD221 pKa = 3.33PALRR225 pKa = 11.84EE226 pKa = 4.46CITRR230 pKa = 11.84TCRR233 pKa = 11.84EE234 pKa = 3.67IFGGMKK240 pKa = 10.03FPEE243 pKa = 4.38TTPVLTQNVSSKK255 pKa = 11.18SSLHH259 pKa = 5.58SNRR262 pKa = 11.84SEE264 pKa = 3.83GGALSEE270 pKa = 4.53IFSEE274 pKa = 4.28EE275 pKa = 4.55DD276 pKa = 3.02PLEE279 pKa = 4.25YY280 pKa = 10.68YY281 pKa = 10.51GEE283 pKa = 4.57CFTEE287 pKa = 4.04EE288 pKa = 4.08TGPLSIKK295 pKa = 8.02TNKK298 pKa = 8.27FAAMEE303 pKa = 3.96KK304 pKa = 10.02LGQIRR309 pKa = 11.84EE310 pKa = 3.94RR311 pKa = 11.84TARR314 pKa = 11.84VLRR317 pKa = 11.84SDD319 pKa = 3.94EE320 pKa = 4.09GSKK323 pKa = 10.78LSSHH327 pKa = 6.15WQTQAILEE335 pKa = 4.1PLKK338 pKa = 10.92CRR340 pKa = 11.84MITKK344 pKa = 10.67GEE346 pKa = 4.09LRR348 pKa = 11.84WNAAWSDD355 pKa = 3.26IQKK358 pKa = 10.69EE359 pKa = 4.12MFRR362 pKa = 11.84RR363 pKa = 11.84LGRR366 pKa = 11.84YY367 pKa = 7.39EE368 pKa = 3.87QFKK371 pKa = 9.07LTRR374 pKa = 11.84GHH376 pKa = 6.78EE377 pKa = 4.0IEE379 pKa = 4.71KK380 pKa = 10.16QDD382 pKa = 3.43SLMFQVSGDD391 pKa = 3.95PLDD394 pKa = 4.93LDD396 pKa = 4.02GDD398 pKa = 3.92QFYY401 pKa = 11.39VAGDD405 pKa = 3.74FSAATDD411 pKa = 3.79EE412 pKa = 4.68LKK414 pKa = 10.92ADD416 pKa = 3.62ATRR419 pKa = 11.84AAIEE423 pKa = 4.47GFTDD427 pKa = 3.6PVLRR431 pKa = 11.84NLLTVNLLNGLINYY445 pKa = 7.01PPKK448 pKa = 10.69YY449 pKa = 10.07EE450 pKa = 4.32IDD452 pKa = 3.71PVHH455 pKa = 5.81QANGQLMGSIFSFPLLCAINLAIYY479 pKa = 9.93RR480 pKa = 11.84YY481 pKa = 6.88TWEE484 pKa = 4.36SRR486 pKa = 11.84DD487 pKa = 3.22KK488 pKa = 11.38SGFRR492 pKa = 11.84KK493 pKa = 10.01PIDD496 pKa = 3.67SLPVLINGDD505 pKa = 4.18DD506 pKa = 3.2IAFRR510 pKa = 11.84TTRR513 pKa = 11.84WFYY516 pKa = 11.48EE517 pKa = 3.92DD518 pKa = 2.9WKK520 pKa = 10.93KK521 pKa = 11.15NSAEE525 pKa = 4.03AGLIASRR532 pKa = 11.84GKK534 pKa = 10.84CYY536 pKa = 8.38FTKK539 pKa = 10.55KK540 pKa = 10.2FFLVNSRR547 pKa = 11.84LIHH550 pKa = 5.02STPTGILTQAIPYY563 pKa = 9.21VNYY566 pKa = 10.88GLVTGQKK573 pKa = 9.95KK574 pKa = 10.65GEE576 pKa = 4.26GDD578 pKa = 3.62DD579 pKa = 3.9DD580 pKa = 4.07LARR583 pKa = 11.84SRR585 pKa = 11.84RR586 pKa = 11.84EE587 pKa = 3.54KK588 pKa = 10.53LACLEE593 pKa = 4.01GALRR597 pKa = 11.84DD598 pKa = 3.66LSKK601 pKa = 11.41DD602 pKa = 2.91FDD604 pKa = 3.94EE605 pKa = 5.64RR606 pKa = 11.84SEE608 pKa = 3.93ILKK611 pKa = 10.38RR612 pKa = 11.84ATARR616 pKa = 11.84AMKK619 pKa = 10.2RR620 pKa = 11.84GDD622 pKa = 3.18IRR624 pKa = 11.84ISGISKK630 pKa = 8.33TQLGLPSVEE639 pKa = 4.4IDD641 pKa = 3.21DD642 pKa = 5.19DD643 pKa = 3.73EE644 pKa = 4.72ARR646 pKa = 11.84RR647 pKa = 11.84YY648 pKa = 7.85LTYY651 pKa = 10.83ASRR654 pKa = 11.84IRR656 pKa = 11.84DD657 pKa = 3.54RR658 pKa = 11.84GQKK661 pKa = 9.64EE662 pKa = 3.73PSAGHH667 pKa = 6.16TGLAPFIFKK676 pKa = 10.31FPRR679 pKa = 11.84VKK681 pKa = 10.17TFGNLRR687 pKa = 11.84TKK689 pKa = 7.39QHH691 pKa = 6.35KK692 pKa = 10.3AVDD695 pKa = 4.31LVKK698 pKa = 10.23HH699 pKa = 5.93LAMVGEE705 pKa = 4.46KK706 pKa = 9.93KK707 pKa = 9.94EE708 pKa = 3.77KK709 pKa = 9.73RR710 pKa = 11.84ARR712 pKa = 11.84EE713 pKa = 3.93EE714 pKa = 3.71EE715 pKa = 3.94ALMEE719 pKa = 4.16VLSAQRR725 pKa = 11.84KK726 pKa = 8.74RR727 pKa = 11.84GFAFDD732 pKa = 3.38IGRR735 pKa = 11.84LPEE738 pKa = 4.12SSEE741 pKa = 3.86EE742 pKa = 3.44ILYY745 pKa = 10.12NKK747 pKa = 8.89KK748 pKa = 10.39RR749 pKa = 11.84KK750 pKa = 9.17
MM1 pKa = 7.48SKK3 pKa = 10.62SDD5 pKa = 3.94DD6 pKa = 3.69RR7 pKa = 11.84RR8 pKa = 11.84PSDD11 pKa = 3.02SQYY14 pKa = 11.06PLRR17 pKa = 11.84LSVSKK22 pKa = 9.82TKK24 pKa = 10.67DD25 pKa = 2.93EE26 pKa = 3.92EE27 pKa = 4.15AFRR30 pKa = 11.84VGLLQEE36 pKa = 3.94PRR38 pKa = 11.84CRR40 pKa = 11.84KK41 pKa = 9.3VALLLAPSCKK51 pKa = 9.74VEE53 pKa = 4.52SVATTSPADD62 pKa = 3.58PLKK65 pKa = 10.96PPGADD70 pKa = 2.7NWRR73 pKa = 11.84LSPSDD78 pKa = 3.55GQIVTTLKK86 pKa = 10.03IVSIITAAIEE96 pKa = 4.18DD97 pKa = 3.68VHH99 pKa = 7.73DD100 pKa = 3.6FHH102 pKa = 9.26LNDD105 pKa = 3.63RR106 pKa = 11.84QIVQAVLMWLPYY118 pKa = 10.9VMADD122 pKa = 2.66VWGPVLKK129 pKa = 10.49ARR131 pKa = 11.84KK132 pKa = 9.58DD133 pKa = 3.3ILFAYY138 pKa = 10.1ALRR141 pKa = 11.84QEE143 pKa = 5.07EE144 pKa = 5.46IPPPWCEE151 pKa = 4.25CCNQTYY157 pKa = 10.41ILPGRR162 pKa = 11.84AGSALRR168 pKa = 11.84QRR170 pKa = 11.84VLYY173 pKa = 10.31NKK175 pKa = 10.33NEE177 pKa = 4.31DD178 pKa = 3.26NRR180 pKa = 11.84IFANTILHH188 pKa = 5.82GLKK191 pKa = 10.27KK192 pKa = 10.58GMPKK196 pKa = 10.31LSEE199 pKa = 3.7AHH201 pKa = 6.85VIDD204 pKa = 5.49GYY206 pKa = 10.87LKK208 pKa = 10.07HH209 pKa = 6.9RR210 pKa = 11.84EE211 pKa = 3.96RR212 pKa = 11.84LSTDD216 pKa = 2.92KK217 pKa = 11.5GPVDD221 pKa = 3.33PALRR225 pKa = 11.84EE226 pKa = 4.46CITRR230 pKa = 11.84TCRR233 pKa = 11.84EE234 pKa = 3.67IFGGMKK240 pKa = 10.03FPEE243 pKa = 4.38TTPVLTQNVSSKK255 pKa = 11.18SSLHH259 pKa = 5.58SNRR262 pKa = 11.84SEE264 pKa = 3.83GGALSEE270 pKa = 4.53IFSEE274 pKa = 4.28EE275 pKa = 4.55DD276 pKa = 3.02PLEE279 pKa = 4.25YY280 pKa = 10.68YY281 pKa = 10.51GEE283 pKa = 4.57CFTEE287 pKa = 4.04EE288 pKa = 4.08TGPLSIKK295 pKa = 8.02TNKK298 pKa = 8.27FAAMEE303 pKa = 3.96KK304 pKa = 10.02LGQIRR309 pKa = 11.84EE310 pKa = 3.94RR311 pKa = 11.84TARR314 pKa = 11.84VLRR317 pKa = 11.84SDD319 pKa = 3.94EE320 pKa = 4.09GSKK323 pKa = 10.78LSSHH327 pKa = 6.15WQTQAILEE335 pKa = 4.1PLKK338 pKa = 10.92CRR340 pKa = 11.84MITKK344 pKa = 10.67GEE346 pKa = 4.09LRR348 pKa = 11.84WNAAWSDD355 pKa = 3.26IQKK358 pKa = 10.69EE359 pKa = 4.12MFRR362 pKa = 11.84RR363 pKa = 11.84LGRR366 pKa = 11.84YY367 pKa = 7.39EE368 pKa = 3.87QFKK371 pKa = 9.07LTRR374 pKa = 11.84GHH376 pKa = 6.78EE377 pKa = 4.0IEE379 pKa = 4.71KK380 pKa = 10.16QDD382 pKa = 3.43SLMFQVSGDD391 pKa = 3.95PLDD394 pKa = 4.93LDD396 pKa = 4.02GDD398 pKa = 3.92QFYY401 pKa = 11.39VAGDD405 pKa = 3.74FSAATDD411 pKa = 3.79EE412 pKa = 4.68LKK414 pKa = 10.92ADD416 pKa = 3.62ATRR419 pKa = 11.84AAIEE423 pKa = 4.47GFTDD427 pKa = 3.6PVLRR431 pKa = 11.84NLLTVNLLNGLINYY445 pKa = 7.01PPKK448 pKa = 10.69YY449 pKa = 10.07EE450 pKa = 4.32IDD452 pKa = 3.71PVHH455 pKa = 5.81QANGQLMGSIFSFPLLCAINLAIYY479 pKa = 9.93RR480 pKa = 11.84YY481 pKa = 6.88TWEE484 pKa = 4.36SRR486 pKa = 11.84DD487 pKa = 3.22KK488 pKa = 11.38SGFRR492 pKa = 11.84KK493 pKa = 10.01PIDD496 pKa = 3.67SLPVLINGDD505 pKa = 4.18DD506 pKa = 3.2IAFRR510 pKa = 11.84TTRR513 pKa = 11.84WFYY516 pKa = 11.48EE517 pKa = 3.92DD518 pKa = 2.9WKK520 pKa = 10.93KK521 pKa = 11.15NSAEE525 pKa = 4.03AGLIASRR532 pKa = 11.84GKK534 pKa = 10.84CYY536 pKa = 8.38FTKK539 pKa = 10.55KK540 pKa = 10.2FFLVNSRR547 pKa = 11.84LIHH550 pKa = 5.02STPTGILTQAIPYY563 pKa = 9.21VNYY566 pKa = 10.88GLVTGQKK573 pKa = 9.95KK574 pKa = 10.65GEE576 pKa = 4.26GDD578 pKa = 3.62DD579 pKa = 3.9DD580 pKa = 4.07LARR583 pKa = 11.84SRR585 pKa = 11.84RR586 pKa = 11.84EE587 pKa = 3.54KK588 pKa = 10.53LACLEE593 pKa = 4.01GALRR597 pKa = 11.84DD598 pKa = 3.66LSKK601 pKa = 11.41DD602 pKa = 2.91FDD604 pKa = 3.94EE605 pKa = 5.64RR606 pKa = 11.84SEE608 pKa = 3.93ILKK611 pKa = 10.38RR612 pKa = 11.84ATARR616 pKa = 11.84AMKK619 pKa = 10.2RR620 pKa = 11.84GDD622 pKa = 3.18IRR624 pKa = 11.84ISGISKK630 pKa = 8.33TQLGLPSVEE639 pKa = 4.4IDD641 pKa = 3.21DD642 pKa = 5.19DD643 pKa = 3.73EE644 pKa = 4.72ARR646 pKa = 11.84RR647 pKa = 11.84YY648 pKa = 7.85LTYY651 pKa = 10.83ASRR654 pKa = 11.84IRR656 pKa = 11.84DD657 pKa = 3.54RR658 pKa = 11.84GQKK661 pKa = 9.64EE662 pKa = 3.73PSAGHH667 pKa = 6.16TGLAPFIFKK676 pKa = 10.31FPRR679 pKa = 11.84VKK681 pKa = 10.17TFGNLRR687 pKa = 11.84TKK689 pKa = 7.39QHH691 pKa = 6.35KK692 pKa = 10.3AVDD695 pKa = 4.31LVKK698 pKa = 10.23HH699 pKa = 5.93LAMVGEE705 pKa = 4.46KK706 pKa = 9.93KK707 pKa = 9.94EE708 pKa = 3.77KK709 pKa = 9.73RR710 pKa = 11.84ARR712 pKa = 11.84EE713 pKa = 3.93EE714 pKa = 3.71EE715 pKa = 3.94ALMEE719 pKa = 4.16VLSAQRR725 pKa = 11.84KK726 pKa = 8.74RR727 pKa = 11.84GFAFDD732 pKa = 3.38IGRR735 pKa = 11.84LPEE738 pKa = 4.12SSEE741 pKa = 3.86EE742 pKa = 3.44ILYY745 pKa = 10.12NKK747 pKa = 8.89KK748 pKa = 10.39RR749 pKa = 11.84KK750 pKa = 9.17
Molecular weight: 84.95 kDa
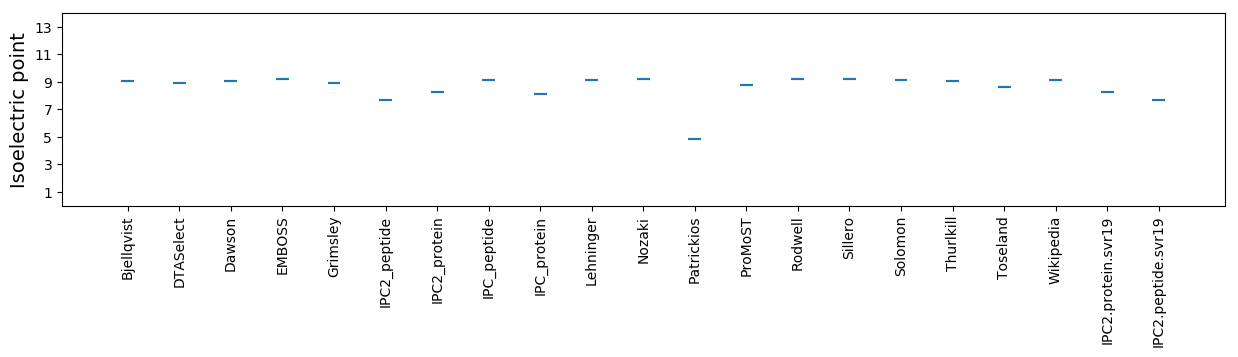
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIB0|A0A1L3KIB0_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 12 OX=1922439 PE=4 SV=1
MM1 pKa = 7.48SKK3 pKa = 10.62SDD5 pKa = 3.94DD6 pKa = 3.69RR7 pKa = 11.84RR8 pKa = 11.84PSDD11 pKa = 3.02SQYY14 pKa = 11.06PLRR17 pKa = 11.84LSVSKK22 pKa = 9.82TKK24 pKa = 10.67DD25 pKa = 2.93EE26 pKa = 3.92EE27 pKa = 4.15AFRR30 pKa = 11.84VGLLQEE36 pKa = 3.94PRR38 pKa = 11.84CRR40 pKa = 11.84KK41 pKa = 9.3VALLLAPSCKK51 pKa = 9.74VEE53 pKa = 4.52SVATTSPADD62 pKa = 3.58PLKK65 pKa = 10.96PPGADD70 pKa = 2.7NWRR73 pKa = 11.84LSPSDD78 pKa = 3.55GQIVTTLKK86 pKa = 10.03IVSIITAAIEE96 pKa = 4.18DD97 pKa = 3.68VHH99 pKa = 7.73DD100 pKa = 3.6FHH102 pKa = 9.26LNDD105 pKa = 3.63RR106 pKa = 11.84QIVQAVLMWLPYY118 pKa = 10.9VMADD122 pKa = 2.66VWGPVLKK129 pKa = 10.49ARR131 pKa = 11.84KK132 pKa = 9.58DD133 pKa = 3.3ILFAYY138 pKa = 10.1ALRR141 pKa = 11.84QEE143 pKa = 5.07EE144 pKa = 5.46IPPPWCEE151 pKa = 4.25CCNQTYY157 pKa = 10.41ILPGRR162 pKa = 11.84AGSALRR168 pKa = 11.84QRR170 pKa = 11.84VLYY173 pKa = 10.31NKK175 pKa = 10.33NEE177 pKa = 4.31DD178 pKa = 3.26NRR180 pKa = 11.84IFANTILHH188 pKa = 5.82GLKK191 pKa = 10.27KK192 pKa = 10.58GMPKK196 pKa = 10.31LSEE199 pKa = 3.7AHH201 pKa = 6.85VIDD204 pKa = 5.49GYY206 pKa = 10.87LKK208 pKa = 10.07HH209 pKa = 6.9RR210 pKa = 11.84EE211 pKa = 3.96RR212 pKa = 11.84LSTDD216 pKa = 2.92KK217 pKa = 11.5GPVDD221 pKa = 3.33PALRR225 pKa = 11.84EE226 pKa = 4.46CITRR230 pKa = 11.84TCRR233 pKa = 11.84EE234 pKa = 3.67IFGGMKK240 pKa = 10.03FPEE243 pKa = 4.38TTPVLTQNVSSKK255 pKa = 11.18SSLHH259 pKa = 5.58SNRR262 pKa = 11.84SEE264 pKa = 3.83GGALSEE270 pKa = 4.53IFSEE274 pKa = 4.28EE275 pKa = 4.55DD276 pKa = 3.02PLEE279 pKa = 4.25YY280 pKa = 10.68YY281 pKa = 10.51GEE283 pKa = 4.57CFTEE287 pKa = 4.04EE288 pKa = 4.08TGPLSIKK295 pKa = 8.02TNKK298 pKa = 8.27FAAMEE303 pKa = 3.96KK304 pKa = 10.02LGQIRR309 pKa = 11.84EE310 pKa = 3.94RR311 pKa = 11.84TARR314 pKa = 11.84VLRR317 pKa = 11.84SDD319 pKa = 3.94EE320 pKa = 4.09GSKK323 pKa = 10.78LSSHH327 pKa = 6.15WQTQAILEE335 pKa = 4.1PLKK338 pKa = 10.92CRR340 pKa = 11.84MITKK344 pKa = 10.67GEE346 pKa = 4.09LRR348 pKa = 11.84WNAAWSDD355 pKa = 3.26IQKK358 pKa = 10.69EE359 pKa = 4.12MFRR362 pKa = 11.84RR363 pKa = 11.84LGRR366 pKa = 11.84YY367 pKa = 7.39EE368 pKa = 3.87QFKK371 pKa = 9.07LTRR374 pKa = 11.84GHH376 pKa = 6.78EE377 pKa = 4.0IEE379 pKa = 4.71KK380 pKa = 10.16QDD382 pKa = 3.43SLMFQVSGDD391 pKa = 3.95PLDD394 pKa = 4.93LDD396 pKa = 4.02GDD398 pKa = 3.92QFYY401 pKa = 11.39VAGDD405 pKa = 3.74FSAATDD411 pKa = 3.79EE412 pKa = 4.68LKK414 pKa = 10.92ADD416 pKa = 3.62ATRR419 pKa = 11.84AAIEE423 pKa = 4.47GFTDD427 pKa = 3.6PVLRR431 pKa = 11.84NLLTVNLLNGLINYY445 pKa = 7.01PPKK448 pKa = 10.69YY449 pKa = 10.07EE450 pKa = 4.32IDD452 pKa = 3.71PVHH455 pKa = 5.81QANGQLMGSIFSFPLLCAINLAIYY479 pKa = 9.93RR480 pKa = 11.84YY481 pKa = 6.88TWEE484 pKa = 4.36SRR486 pKa = 11.84DD487 pKa = 3.22KK488 pKa = 11.38SGFRR492 pKa = 11.84KK493 pKa = 10.01PIDD496 pKa = 3.67SLPVLINGDD505 pKa = 4.18DD506 pKa = 3.2IAFRR510 pKa = 11.84TTRR513 pKa = 11.84WFYY516 pKa = 11.48EE517 pKa = 3.92DD518 pKa = 2.9WKK520 pKa = 10.93KK521 pKa = 11.15NSAEE525 pKa = 4.03AGLIASRR532 pKa = 11.84GKK534 pKa = 10.84CYY536 pKa = 8.38FTKK539 pKa = 10.55KK540 pKa = 10.2FFLVNSRR547 pKa = 11.84LIHH550 pKa = 5.02STPTGILTQAIPYY563 pKa = 9.21VNYY566 pKa = 10.88GLVTGQKK573 pKa = 9.95KK574 pKa = 10.65GEE576 pKa = 4.26GDD578 pKa = 3.62DD579 pKa = 3.9DD580 pKa = 4.07LARR583 pKa = 11.84SRR585 pKa = 11.84RR586 pKa = 11.84EE587 pKa = 3.54KK588 pKa = 10.53LACLEE593 pKa = 4.01GALRR597 pKa = 11.84DD598 pKa = 3.66LSKK601 pKa = 11.41DD602 pKa = 2.91FDD604 pKa = 3.94EE605 pKa = 5.64RR606 pKa = 11.84SEE608 pKa = 3.93ILKK611 pKa = 10.38RR612 pKa = 11.84ATARR616 pKa = 11.84AMKK619 pKa = 10.2RR620 pKa = 11.84GDD622 pKa = 3.18IRR624 pKa = 11.84ISGISKK630 pKa = 8.33TQLGLPSVEE639 pKa = 4.4IDD641 pKa = 3.21DD642 pKa = 5.19DD643 pKa = 3.73EE644 pKa = 4.72ARR646 pKa = 11.84RR647 pKa = 11.84YY648 pKa = 7.85LTYY651 pKa = 10.83ASRR654 pKa = 11.84IRR656 pKa = 11.84DD657 pKa = 3.54RR658 pKa = 11.84GQKK661 pKa = 9.64EE662 pKa = 3.73PSAGHH667 pKa = 6.16TGLAPFIFKK676 pKa = 10.31FPRR679 pKa = 11.84VKK681 pKa = 10.17TFGNLRR687 pKa = 11.84TKK689 pKa = 7.39QHH691 pKa = 6.35KK692 pKa = 10.3AVDD695 pKa = 4.31LVKK698 pKa = 10.23HH699 pKa = 5.93LAMVGEE705 pKa = 4.46KK706 pKa = 9.93KK707 pKa = 9.94EE708 pKa = 3.77KK709 pKa = 9.73RR710 pKa = 11.84ARR712 pKa = 11.84EE713 pKa = 3.93EE714 pKa = 3.71EE715 pKa = 3.94ALMEE719 pKa = 4.16VLSAQRR725 pKa = 11.84KK726 pKa = 8.74RR727 pKa = 11.84GFAFDD732 pKa = 3.38IGRR735 pKa = 11.84LPEE738 pKa = 4.12SSEE741 pKa = 3.86EE742 pKa = 3.44ILYY745 pKa = 10.12NKK747 pKa = 8.89KK748 pKa = 10.39RR749 pKa = 11.84KK750 pKa = 9.17
MM1 pKa = 7.48SKK3 pKa = 10.62SDD5 pKa = 3.94DD6 pKa = 3.69RR7 pKa = 11.84RR8 pKa = 11.84PSDD11 pKa = 3.02SQYY14 pKa = 11.06PLRR17 pKa = 11.84LSVSKK22 pKa = 9.82TKK24 pKa = 10.67DD25 pKa = 2.93EE26 pKa = 3.92EE27 pKa = 4.15AFRR30 pKa = 11.84VGLLQEE36 pKa = 3.94PRR38 pKa = 11.84CRR40 pKa = 11.84KK41 pKa = 9.3VALLLAPSCKK51 pKa = 9.74VEE53 pKa = 4.52SVATTSPADD62 pKa = 3.58PLKK65 pKa = 10.96PPGADD70 pKa = 2.7NWRR73 pKa = 11.84LSPSDD78 pKa = 3.55GQIVTTLKK86 pKa = 10.03IVSIITAAIEE96 pKa = 4.18DD97 pKa = 3.68VHH99 pKa = 7.73DD100 pKa = 3.6FHH102 pKa = 9.26LNDD105 pKa = 3.63RR106 pKa = 11.84QIVQAVLMWLPYY118 pKa = 10.9VMADD122 pKa = 2.66VWGPVLKK129 pKa = 10.49ARR131 pKa = 11.84KK132 pKa = 9.58DD133 pKa = 3.3ILFAYY138 pKa = 10.1ALRR141 pKa = 11.84QEE143 pKa = 5.07EE144 pKa = 5.46IPPPWCEE151 pKa = 4.25CCNQTYY157 pKa = 10.41ILPGRR162 pKa = 11.84AGSALRR168 pKa = 11.84QRR170 pKa = 11.84VLYY173 pKa = 10.31NKK175 pKa = 10.33NEE177 pKa = 4.31DD178 pKa = 3.26NRR180 pKa = 11.84IFANTILHH188 pKa = 5.82GLKK191 pKa = 10.27KK192 pKa = 10.58GMPKK196 pKa = 10.31LSEE199 pKa = 3.7AHH201 pKa = 6.85VIDD204 pKa = 5.49GYY206 pKa = 10.87LKK208 pKa = 10.07HH209 pKa = 6.9RR210 pKa = 11.84EE211 pKa = 3.96RR212 pKa = 11.84LSTDD216 pKa = 2.92KK217 pKa = 11.5GPVDD221 pKa = 3.33PALRR225 pKa = 11.84EE226 pKa = 4.46CITRR230 pKa = 11.84TCRR233 pKa = 11.84EE234 pKa = 3.67IFGGMKK240 pKa = 10.03FPEE243 pKa = 4.38TTPVLTQNVSSKK255 pKa = 11.18SSLHH259 pKa = 5.58SNRR262 pKa = 11.84SEE264 pKa = 3.83GGALSEE270 pKa = 4.53IFSEE274 pKa = 4.28EE275 pKa = 4.55DD276 pKa = 3.02PLEE279 pKa = 4.25YY280 pKa = 10.68YY281 pKa = 10.51GEE283 pKa = 4.57CFTEE287 pKa = 4.04EE288 pKa = 4.08TGPLSIKK295 pKa = 8.02TNKK298 pKa = 8.27FAAMEE303 pKa = 3.96KK304 pKa = 10.02LGQIRR309 pKa = 11.84EE310 pKa = 3.94RR311 pKa = 11.84TARR314 pKa = 11.84VLRR317 pKa = 11.84SDD319 pKa = 3.94EE320 pKa = 4.09GSKK323 pKa = 10.78LSSHH327 pKa = 6.15WQTQAILEE335 pKa = 4.1PLKK338 pKa = 10.92CRR340 pKa = 11.84MITKK344 pKa = 10.67GEE346 pKa = 4.09LRR348 pKa = 11.84WNAAWSDD355 pKa = 3.26IQKK358 pKa = 10.69EE359 pKa = 4.12MFRR362 pKa = 11.84RR363 pKa = 11.84LGRR366 pKa = 11.84YY367 pKa = 7.39EE368 pKa = 3.87QFKK371 pKa = 9.07LTRR374 pKa = 11.84GHH376 pKa = 6.78EE377 pKa = 4.0IEE379 pKa = 4.71KK380 pKa = 10.16QDD382 pKa = 3.43SLMFQVSGDD391 pKa = 3.95PLDD394 pKa = 4.93LDD396 pKa = 4.02GDD398 pKa = 3.92QFYY401 pKa = 11.39VAGDD405 pKa = 3.74FSAATDD411 pKa = 3.79EE412 pKa = 4.68LKK414 pKa = 10.92ADD416 pKa = 3.62ATRR419 pKa = 11.84AAIEE423 pKa = 4.47GFTDD427 pKa = 3.6PVLRR431 pKa = 11.84NLLTVNLLNGLINYY445 pKa = 7.01PPKK448 pKa = 10.69YY449 pKa = 10.07EE450 pKa = 4.32IDD452 pKa = 3.71PVHH455 pKa = 5.81QANGQLMGSIFSFPLLCAINLAIYY479 pKa = 9.93RR480 pKa = 11.84YY481 pKa = 6.88TWEE484 pKa = 4.36SRR486 pKa = 11.84DD487 pKa = 3.22KK488 pKa = 11.38SGFRR492 pKa = 11.84KK493 pKa = 10.01PIDD496 pKa = 3.67SLPVLINGDD505 pKa = 4.18DD506 pKa = 3.2IAFRR510 pKa = 11.84TTRR513 pKa = 11.84WFYY516 pKa = 11.48EE517 pKa = 3.92DD518 pKa = 2.9WKK520 pKa = 10.93KK521 pKa = 11.15NSAEE525 pKa = 4.03AGLIASRR532 pKa = 11.84GKK534 pKa = 10.84CYY536 pKa = 8.38FTKK539 pKa = 10.55KK540 pKa = 10.2FFLVNSRR547 pKa = 11.84LIHH550 pKa = 5.02STPTGILTQAIPYY563 pKa = 9.21VNYY566 pKa = 10.88GLVTGQKK573 pKa = 9.95KK574 pKa = 10.65GEE576 pKa = 4.26GDD578 pKa = 3.62DD579 pKa = 3.9DD580 pKa = 4.07LARR583 pKa = 11.84SRR585 pKa = 11.84RR586 pKa = 11.84EE587 pKa = 3.54KK588 pKa = 10.53LACLEE593 pKa = 4.01GALRR597 pKa = 11.84DD598 pKa = 3.66LSKK601 pKa = 11.41DD602 pKa = 2.91FDD604 pKa = 3.94EE605 pKa = 5.64RR606 pKa = 11.84SEE608 pKa = 3.93ILKK611 pKa = 10.38RR612 pKa = 11.84ATARR616 pKa = 11.84AMKK619 pKa = 10.2RR620 pKa = 11.84GDD622 pKa = 3.18IRR624 pKa = 11.84ISGISKK630 pKa = 8.33TQLGLPSVEE639 pKa = 4.4IDD641 pKa = 3.21DD642 pKa = 5.19DD643 pKa = 3.73EE644 pKa = 4.72ARR646 pKa = 11.84RR647 pKa = 11.84YY648 pKa = 7.85LTYY651 pKa = 10.83ASRR654 pKa = 11.84IRR656 pKa = 11.84DD657 pKa = 3.54RR658 pKa = 11.84GQKK661 pKa = 9.64EE662 pKa = 3.73PSAGHH667 pKa = 6.16TGLAPFIFKK676 pKa = 10.31FPRR679 pKa = 11.84VKK681 pKa = 10.17TFGNLRR687 pKa = 11.84TKK689 pKa = 7.39QHH691 pKa = 6.35KK692 pKa = 10.3AVDD695 pKa = 4.31LVKK698 pKa = 10.23HH699 pKa = 5.93LAMVGEE705 pKa = 4.46KK706 pKa = 9.93KK707 pKa = 9.94EE708 pKa = 3.77KK709 pKa = 9.73RR710 pKa = 11.84ARR712 pKa = 11.84EE713 pKa = 3.93EE714 pKa = 3.71EE715 pKa = 3.94ALMEE719 pKa = 4.16VLSAQRR725 pKa = 11.84KK726 pKa = 8.74RR727 pKa = 11.84GFAFDD732 pKa = 3.38IGRR735 pKa = 11.84LPEE738 pKa = 4.12SSEE741 pKa = 3.86EE742 pKa = 3.44ILYY745 pKa = 10.12NKK747 pKa = 8.89KK748 pKa = 10.39RR749 pKa = 11.84KK750 pKa = 9.17
Molecular weight: 84.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
750 |
750 |
750 |
750.0 |
84.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.467 ± 0.0 | 1.6 ± 0.0 |
6.267 ± 0.0 | 7.2 ± 0.0 |
4.0 ± 0.0 | 6.4 ± 0.0 |
1.733 ± 0.0 | 5.733 ± 0.0 |
7.333 ± 0.0 | 10.133 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.733 ± 0.0 | 3.067 ± 0.0 |
5.067 ± 0.0 | 3.333 ± 0.0 |
8.133 ± 0.0 | 6.933 ± 0.0 |
5.2 ± 0.0 | 4.533 ± 0.0 |
1.333 ± 0.0 | 2.8 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
