
Prolixibacter denitrificans
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Prolixibacter
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
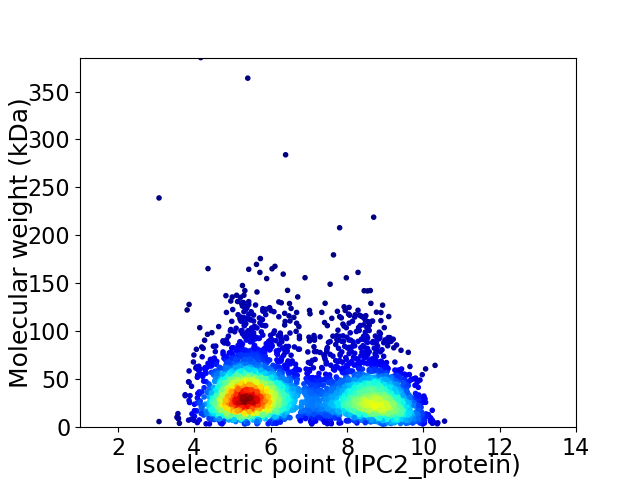
Virtual 2D-PAGE plot for 3987 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
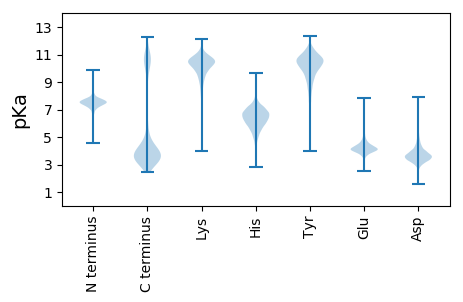
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P8CFW4|A0A2P8CFW4_9BACT MoxR-like ATPase OS=Prolixibacter denitrificans OX=1541063 GN=CLV93_103260 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.56NIFTPLFLILLITIFSNKK20 pKa = 8.94SFAQTGPPTPPDD32 pKa = 4.17PDD34 pKa = 4.28SNDD37 pKa = 3.4TEE39 pKa = 4.21NALFDD44 pKa = 4.29NDD46 pKa = 5.35PIPEE50 pKa = 4.34FGSVTFSPSPMTDD63 pKa = 2.88TEE65 pKa = 4.48VTGNNTISASISDD78 pKa = 4.13DD79 pKa = 3.83GEE81 pKa = 4.37VISALVLYY89 pKa = 7.41GTTSGDD95 pKa = 3.39LTKK98 pKa = 10.89SFALSYY104 pKa = 11.16SSGTTWKK111 pKa = 10.63ADD113 pKa = 3.56LPVLEE118 pKa = 5.79DD119 pKa = 3.06GTYY122 pKa = 10.07YY123 pKa = 10.71YY124 pKa = 10.21QIKK127 pKa = 10.19AADD130 pKa = 3.6NSQQEE135 pKa = 4.64VVSTEE140 pKa = 3.5ASFEE144 pKa = 4.13ITAGTPVAPADD155 pKa = 4.07LQASAIDD162 pKa = 3.77YY163 pKa = 11.09NRR165 pKa = 11.84FTLEE169 pKa = 3.84WQPSYY174 pKa = 11.02QATSYY179 pKa = 10.96YY180 pKa = 10.53LIISTEE186 pKa = 3.85EE187 pKa = 4.46DD188 pKa = 3.77YY189 pKa = 11.76SNPIVDD195 pKa = 3.83GDD197 pKa = 3.61IGNVTSYY204 pKa = 10.71EE205 pKa = 4.0ATSVQFSTTYY215 pKa = 10.46YY216 pKa = 9.09YY217 pKa = 10.17QLKK220 pKa = 10.28AIRR223 pKa = 11.84LMDD226 pKa = 5.23DD227 pKa = 3.46DD228 pKa = 5.65AVMEE232 pKa = 4.46SEE234 pKa = 3.99ISSGEE239 pKa = 4.22VTTLDD244 pKa = 3.31QPAPEE249 pKa = 4.16VTVSVEE255 pKa = 4.0SLPDD259 pKa = 3.81FGDD262 pKa = 3.62VVSGEE267 pKa = 4.1NSATSSYY274 pKa = 10.8TVTAQYY280 pKa = 11.12LRR282 pKa = 11.84DD283 pKa = 3.98NLVVTPPTGFEE294 pKa = 4.03VSTDD298 pKa = 3.31EE299 pKa = 5.28TNWQGNASPLSLAGTDD315 pKa = 4.26GEE317 pKa = 4.74LSTTIYY323 pKa = 10.86VRR325 pKa = 11.84FAPDD329 pKa = 3.25TPGATSGDD337 pKa = 3.42ITHH340 pKa = 6.58EE341 pKa = 4.23TTEE344 pKa = 4.1LPEE347 pKa = 4.28TVLVALSGTALEE359 pKa = 4.54TEE361 pKa = 4.55PANHH365 pKa = 5.96VSNFRR370 pKa = 11.84LAGQNGNTFHH380 pKa = 7.53LSWDD384 pKa = 3.71EE385 pKa = 4.13NNGEE389 pKa = 4.07EE390 pKa = 4.42AAEE393 pKa = 4.99GYY395 pKa = 10.79LLLFTQTTVQTPVDD409 pKa = 4.06GEE411 pKa = 4.56TPIEE415 pKa = 5.26DD416 pKa = 3.37IDD418 pKa = 4.4FSDD421 pKa = 4.55GEE423 pKa = 4.17ALIVIDD429 pKa = 5.02KK430 pKa = 8.31DD431 pKa = 3.39TTAAEE436 pKa = 4.45VYY438 pKa = 10.26IPGSEE443 pKa = 4.21TRR445 pKa = 11.84WYY447 pKa = 8.1TQIIPYY453 pKa = 9.77RR454 pKa = 11.84GSDD457 pKa = 2.87EE458 pKa = 4.29TTNVKK463 pKa = 9.43TDD465 pKa = 3.29GTVPEE470 pKa = 4.41ITLITPLDD478 pKa = 4.33DD479 pKa = 6.28LPNAWINEE487 pKa = 3.73FHH489 pKa = 6.97YY490 pKa = 11.19SDD492 pKa = 5.75DD493 pKa = 4.38GDD495 pKa = 3.88DD496 pKa = 3.69HH497 pKa = 7.34AQVEE501 pKa = 4.8IVLEE505 pKa = 4.21NADD508 pKa = 4.04NYY510 pKa = 11.15SLEE513 pKa = 4.18DD514 pKa = 4.51FILIHH519 pKa = 6.21YY520 pKa = 8.77NGAGGTIIDD529 pKa = 4.39EE530 pKa = 4.72FNFADD535 pKa = 3.71NASVSVQGAFSIILIDD551 pKa = 5.06CGTLQKK557 pKa = 11.14GPDD560 pKa = 3.79GLAISYY566 pKa = 7.19QHH568 pKa = 7.12RR569 pKa = 11.84IVQFISYY576 pKa = 9.3EE577 pKa = 3.9DD578 pKa = 3.55SFIATEE584 pKa = 4.36GDD586 pKa = 3.14ATGITSIEE594 pKa = 4.07IAPQEE599 pKa = 4.46TTTTPPNFSIQLTGSGSYY617 pKa = 10.67YY618 pKa = 10.68EE619 pKa = 5.04DD620 pKa = 4.13FTWQDD625 pKa = 3.32PAEE628 pKa = 4.33SSFGAVNAGQILTASPNLEE647 pKa = 4.33TTWLGTQDD655 pKa = 4.35NDD657 pKa = 3.69WNNAANWNNGVPDD670 pKa = 3.83EE671 pKa = 4.34TMKK674 pKa = 11.27GIIPAGVDD682 pKa = 3.29HH683 pKa = 6.75YY684 pKa = 11.0PVLTAGVTPKK694 pKa = 10.27TLVLEE699 pKa = 4.62SDD701 pKa = 3.79ASLSGQQYY709 pKa = 10.53LPEE712 pKa = 4.32SCILYY717 pKa = 10.48AKK719 pKa = 10.28RR720 pKa = 11.84ALTAEE725 pKa = 3.94KK726 pKa = 9.86WLFLTPPVEE735 pKa = 4.4DD736 pKa = 4.61NIAAMFEE743 pKa = 4.19PVYY746 pKa = 11.2GMVYY750 pKa = 10.57LITYY754 pKa = 9.04DD755 pKa = 3.28NSQAADD761 pKa = 3.6EE762 pKa = 4.62SAWNYY767 pKa = 10.46VSDD770 pKa = 3.7QTLPLSPMTGYY781 pKa = 11.02GFYY784 pKa = 9.56STRR787 pKa = 11.84EE788 pKa = 3.79EE789 pKa = 4.47LEE791 pKa = 4.09LVFNGSPISGTQSVLLSFEE810 pKa = 4.27DD811 pKa = 3.6AGNNFNFVGNPGLGAIDD828 pKa = 3.64WSTSNQANTTGTAYY842 pKa = 10.3MFDD845 pKa = 4.42PNAKK849 pKa = 8.94TYY851 pKa = 9.25LTIASDD857 pKa = 3.93GTVTNPEE864 pKa = 4.32FTDD867 pKa = 4.62MIPPMQGFFVEE878 pKa = 4.43ATNAGNYY885 pKa = 9.19SVNLDD890 pKa = 3.61DD891 pKa = 6.4CVPTDD896 pKa = 4.44QYY898 pKa = 11.53FLKK901 pKa = 10.73QGVPEE906 pKa = 4.41DD907 pKa = 3.72YY908 pKa = 10.34IVRR911 pKa = 11.84LKK913 pKa = 9.51VTGPNGHH920 pKa = 7.12DD921 pKa = 3.33FALVRR926 pKa = 11.84FDD928 pKa = 3.32QTNNDD933 pKa = 3.41EE934 pKa = 4.83LDD936 pKa = 3.68FNEE939 pKa = 4.66DD940 pKa = 2.99SRR942 pKa = 11.84KK943 pKa = 9.71LVYY946 pKa = 10.58YY947 pKa = 10.35DD948 pKa = 3.61IGVPQLWFSQEE959 pKa = 3.98GEE961 pKa = 4.02KK962 pKa = 10.8LAINQARR969 pKa = 11.84TYY971 pKa = 9.44PASFPLGLYY980 pKa = 10.11ISDD983 pKa = 4.43DD984 pKa = 4.04SEE986 pKa = 4.54LSITGTVSGSFSDD999 pKa = 5.19GITMSLEE1006 pKa = 3.71NTITGEE1012 pKa = 4.13TLDD1015 pKa = 4.63LRR1017 pKa = 11.84SSDD1020 pKa = 3.37TLRR1023 pKa = 11.84YY1024 pKa = 9.4DD1025 pKa = 4.05ASAGEE1030 pKa = 4.13NNSLILHH1037 pKa = 6.69LKK1039 pKa = 9.91NGTGFDD1045 pKa = 3.34EE1046 pKa = 4.66FTNEE1050 pKa = 3.4KK1051 pKa = 9.74VRR1053 pKa = 11.84IYY1055 pKa = 10.85SSGEE1059 pKa = 3.71ILHH1062 pKa = 5.6VTASQILKK1070 pKa = 10.66GKK1072 pKa = 10.61LEE1074 pKa = 3.94VFNFIGQKK1082 pKa = 7.17ITEE1085 pKa = 4.0YY1086 pKa = 10.75SINHH1090 pKa = 5.59TNHH1093 pKa = 6.29ARR1095 pKa = 11.84VQPGTGIYY1103 pKa = 9.84LVRR1106 pKa = 11.84WTDD1109 pKa = 3.31DD1110 pKa = 3.3EE1111 pKa = 4.3QAVVQKK1117 pKa = 10.98VSLRR1121 pKa = 3.66
MM1 pKa = 7.44KK2 pKa = 10.56NIFTPLFLILLITIFSNKK20 pKa = 8.94SFAQTGPPTPPDD32 pKa = 4.17PDD34 pKa = 4.28SNDD37 pKa = 3.4TEE39 pKa = 4.21NALFDD44 pKa = 4.29NDD46 pKa = 5.35PIPEE50 pKa = 4.34FGSVTFSPSPMTDD63 pKa = 2.88TEE65 pKa = 4.48VTGNNTISASISDD78 pKa = 4.13DD79 pKa = 3.83GEE81 pKa = 4.37VISALVLYY89 pKa = 7.41GTTSGDD95 pKa = 3.39LTKK98 pKa = 10.89SFALSYY104 pKa = 11.16SSGTTWKK111 pKa = 10.63ADD113 pKa = 3.56LPVLEE118 pKa = 5.79DD119 pKa = 3.06GTYY122 pKa = 10.07YY123 pKa = 10.71YY124 pKa = 10.21QIKK127 pKa = 10.19AADD130 pKa = 3.6NSQQEE135 pKa = 4.64VVSTEE140 pKa = 3.5ASFEE144 pKa = 4.13ITAGTPVAPADD155 pKa = 4.07LQASAIDD162 pKa = 3.77YY163 pKa = 11.09NRR165 pKa = 11.84FTLEE169 pKa = 3.84WQPSYY174 pKa = 11.02QATSYY179 pKa = 10.96YY180 pKa = 10.53LIISTEE186 pKa = 3.85EE187 pKa = 4.46DD188 pKa = 3.77YY189 pKa = 11.76SNPIVDD195 pKa = 3.83GDD197 pKa = 3.61IGNVTSYY204 pKa = 10.71EE205 pKa = 4.0ATSVQFSTTYY215 pKa = 10.46YY216 pKa = 9.09YY217 pKa = 10.17QLKK220 pKa = 10.28AIRR223 pKa = 11.84LMDD226 pKa = 5.23DD227 pKa = 3.46DD228 pKa = 5.65AVMEE232 pKa = 4.46SEE234 pKa = 3.99ISSGEE239 pKa = 4.22VTTLDD244 pKa = 3.31QPAPEE249 pKa = 4.16VTVSVEE255 pKa = 4.0SLPDD259 pKa = 3.81FGDD262 pKa = 3.62VVSGEE267 pKa = 4.1NSATSSYY274 pKa = 10.8TVTAQYY280 pKa = 11.12LRR282 pKa = 11.84DD283 pKa = 3.98NLVVTPPTGFEE294 pKa = 4.03VSTDD298 pKa = 3.31EE299 pKa = 5.28TNWQGNASPLSLAGTDD315 pKa = 4.26GEE317 pKa = 4.74LSTTIYY323 pKa = 10.86VRR325 pKa = 11.84FAPDD329 pKa = 3.25TPGATSGDD337 pKa = 3.42ITHH340 pKa = 6.58EE341 pKa = 4.23TTEE344 pKa = 4.1LPEE347 pKa = 4.28TVLVALSGTALEE359 pKa = 4.54TEE361 pKa = 4.55PANHH365 pKa = 5.96VSNFRR370 pKa = 11.84LAGQNGNTFHH380 pKa = 7.53LSWDD384 pKa = 3.71EE385 pKa = 4.13NNGEE389 pKa = 4.07EE390 pKa = 4.42AAEE393 pKa = 4.99GYY395 pKa = 10.79LLLFTQTTVQTPVDD409 pKa = 4.06GEE411 pKa = 4.56TPIEE415 pKa = 5.26DD416 pKa = 3.37IDD418 pKa = 4.4FSDD421 pKa = 4.55GEE423 pKa = 4.17ALIVIDD429 pKa = 5.02KK430 pKa = 8.31DD431 pKa = 3.39TTAAEE436 pKa = 4.45VYY438 pKa = 10.26IPGSEE443 pKa = 4.21TRR445 pKa = 11.84WYY447 pKa = 8.1TQIIPYY453 pKa = 9.77RR454 pKa = 11.84GSDD457 pKa = 2.87EE458 pKa = 4.29TTNVKK463 pKa = 9.43TDD465 pKa = 3.29GTVPEE470 pKa = 4.41ITLITPLDD478 pKa = 4.33DD479 pKa = 6.28LPNAWINEE487 pKa = 3.73FHH489 pKa = 6.97YY490 pKa = 11.19SDD492 pKa = 5.75DD493 pKa = 4.38GDD495 pKa = 3.88DD496 pKa = 3.69HH497 pKa = 7.34AQVEE501 pKa = 4.8IVLEE505 pKa = 4.21NADD508 pKa = 4.04NYY510 pKa = 11.15SLEE513 pKa = 4.18DD514 pKa = 4.51FILIHH519 pKa = 6.21YY520 pKa = 8.77NGAGGTIIDD529 pKa = 4.39EE530 pKa = 4.72FNFADD535 pKa = 3.71NASVSVQGAFSIILIDD551 pKa = 5.06CGTLQKK557 pKa = 11.14GPDD560 pKa = 3.79GLAISYY566 pKa = 7.19QHH568 pKa = 7.12RR569 pKa = 11.84IVQFISYY576 pKa = 9.3EE577 pKa = 3.9DD578 pKa = 3.55SFIATEE584 pKa = 4.36GDD586 pKa = 3.14ATGITSIEE594 pKa = 4.07IAPQEE599 pKa = 4.46TTTTPPNFSIQLTGSGSYY617 pKa = 10.67YY618 pKa = 10.68EE619 pKa = 5.04DD620 pKa = 4.13FTWQDD625 pKa = 3.32PAEE628 pKa = 4.33SSFGAVNAGQILTASPNLEE647 pKa = 4.33TTWLGTQDD655 pKa = 4.35NDD657 pKa = 3.69WNNAANWNNGVPDD670 pKa = 3.83EE671 pKa = 4.34TMKK674 pKa = 11.27GIIPAGVDD682 pKa = 3.29HH683 pKa = 6.75YY684 pKa = 11.0PVLTAGVTPKK694 pKa = 10.27TLVLEE699 pKa = 4.62SDD701 pKa = 3.79ASLSGQQYY709 pKa = 10.53LPEE712 pKa = 4.32SCILYY717 pKa = 10.48AKK719 pKa = 10.28RR720 pKa = 11.84ALTAEE725 pKa = 3.94KK726 pKa = 9.86WLFLTPPVEE735 pKa = 4.4DD736 pKa = 4.61NIAAMFEE743 pKa = 4.19PVYY746 pKa = 11.2GMVYY750 pKa = 10.57LITYY754 pKa = 9.04DD755 pKa = 3.28NSQAADD761 pKa = 3.6EE762 pKa = 4.62SAWNYY767 pKa = 10.46VSDD770 pKa = 3.7QTLPLSPMTGYY781 pKa = 11.02GFYY784 pKa = 9.56STRR787 pKa = 11.84EE788 pKa = 3.79EE789 pKa = 4.47LEE791 pKa = 4.09LVFNGSPISGTQSVLLSFEE810 pKa = 4.27DD811 pKa = 3.6AGNNFNFVGNPGLGAIDD828 pKa = 3.64WSTSNQANTTGTAYY842 pKa = 10.3MFDD845 pKa = 4.42PNAKK849 pKa = 8.94TYY851 pKa = 9.25LTIASDD857 pKa = 3.93GTVTNPEE864 pKa = 4.32FTDD867 pKa = 4.62MIPPMQGFFVEE878 pKa = 4.43ATNAGNYY885 pKa = 9.19SVNLDD890 pKa = 3.61DD891 pKa = 6.4CVPTDD896 pKa = 4.44QYY898 pKa = 11.53FLKK901 pKa = 10.73QGVPEE906 pKa = 4.41DD907 pKa = 3.72YY908 pKa = 10.34IVRR911 pKa = 11.84LKK913 pKa = 9.51VTGPNGHH920 pKa = 7.12DD921 pKa = 3.33FALVRR926 pKa = 11.84FDD928 pKa = 3.32QTNNDD933 pKa = 3.41EE934 pKa = 4.83LDD936 pKa = 3.68FNEE939 pKa = 4.66DD940 pKa = 2.99SRR942 pKa = 11.84KK943 pKa = 9.71LVYY946 pKa = 10.58YY947 pKa = 10.35DD948 pKa = 3.61IGVPQLWFSQEE959 pKa = 3.98GEE961 pKa = 4.02KK962 pKa = 10.8LAINQARR969 pKa = 11.84TYY971 pKa = 9.44PASFPLGLYY980 pKa = 10.11ISDD983 pKa = 4.43DD984 pKa = 4.04SEE986 pKa = 4.54LSITGTVSGSFSDD999 pKa = 5.19GITMSLEE1006 pKa = 3.71NTITGEE1012 pKa = 4.13TLDD1015 pKa = 4.63LRR1017 pKa = 11.84SSDD1020 pKa = 3.37TLRR1023 pKa = 11.84YY1024 pKa = 9.4DD1025 pKa = 4.05ASAGEE1030 pKa = 4.13NNSLILHH1037 pKa = 6.69LKK1039 pKa = 9.91NGTGFDD1045 pKa = 3.34EE1046 pKa = 4.66FTNEE1050 pKa = 3.4KK1051 pKa = 9.74VRR1053 pKa = 11.84IYY1055 pKa = 10.85SSGEE1059 pKa = 3.71ILHH1062 pKa = 5.6VTASQILKK1070 pKa = 10.66GKK1072 pKa = 10.61LEE1074 pKa = 3.94VFNFIGQKK1082 pKa = 7.17ITEE1085 pKa = 4.0YY1086 pKa = 10.75SINHH1090 pKa = 5.59TNHH1093 pKa = 6.29ARR1095 pKa = 11.84VQPGTGIYY1103 pKa = 9.84LVRR1106 pKa = 11.84WTDD1109 pKa = 3.31DD1110 pKa = 3.3EE1111 pKa = 4.3QAVVQKK1117 pKa = 10.98VSLRR1121 pKa = 3.66
Molecular weight: 122.25 kDa
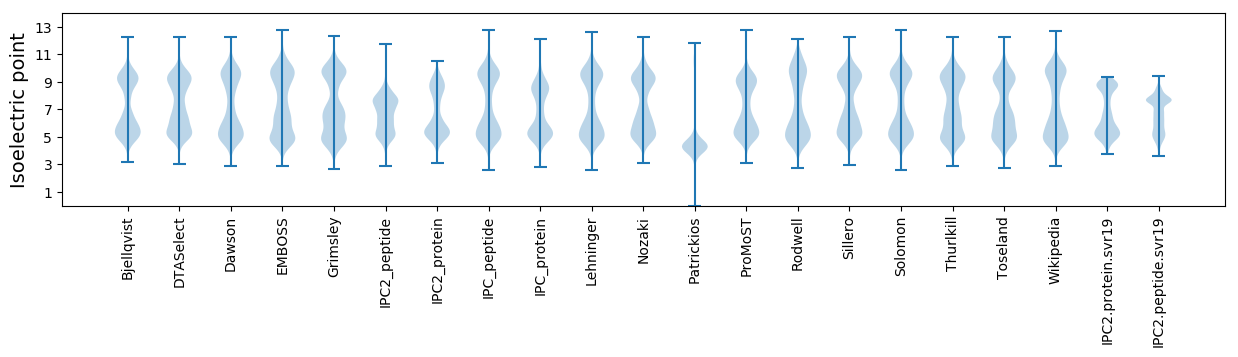
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P8CFC8|A0A2P8CFC8_9BACT CRP-like cAMP-binding protein OS=Prolixibacter denitrificans OX=1541063 GN=CLV93_10393 PE=4 SV=1
MM1 pKa = 7.54SSLGGMTSFFRR12 pKa = 11.84PGMLKK17 pKa = 10.43GALGRR22 pKa = 11.84YY23 pKa = 8.02VFSCATSYY31 pKa = 11.22RR32 pKa = 11.84LDD34 pKa = 3.16ITAIPVKK41 pKa = 9.82WLIKK45 pKa = 8.67STVAIVSHH53 pKa = 6.24SSEE56 pKa = 4.19APSGVWGGYY65 pKa = 6.77SRR67 pKa = 11.84RR68 pKa = 11.84PLRR71 pKa = 11.84EE72 pKa = 3.59LFDD75 pKa = 5.15PNNEE79 pKa = 4.1TPIQLINRR87 pKa = 11.84IARR90 pKa = 11.84RR91 pKa = 11.84ASSILVVVAEE101 pKa = 4.29MSHH104 pKa = 6.62RR105 pKa = 11.84APLLRR110 pKa = 11.84RR111 pKa = 11.84CYY113 pKa = 10.72SPNFFNSHH121 pKa = 4.16QAKK124 pKa = 10.27IPGEE128 pKa = 4.28VPASCDD134 pKa = 3.26EE135 pKa = 4.02NRR137 pKa = 11.84RR138 pKa = 11.84ASPFFLFARR147 pKa = 11.84IGG149 pKa = 3.34
MM1 pKa = 7.54SSLGGMTSFFRR12 pKa = 11.84PGMLKK17 pKa = 10.43GALGRR22 pKa = 11.84YY23 pKa = 8.02VFSCATSYY31 pKa = 11.22RR32 pKa = 11.84LDD34 pKa = 3.16ITAIPVKK41 pKa = 9.82WLIKK45 pKa = 8.67STVAIVSHH53 pKa = 6.24SSEE56 pKa = 4.19APSGVWGGYY65 pKa = 6.77SRR67 pKa = 11.84RR68 pKa = 11.84PLRR71 pKa = 11.84EE72 pKa = 3.59LFDD75 pKa = 5.15PNNEE79 pKa = 4.1TPIQLINRR87 pKa = 11.84IARR90 pKa = 11.84RR91 pKa = 11.84ASSILVVVAEE101 pKa = 4.29MSHH104 pKa = 6.62RR105 pKa = 11.84APLLRR110 pKa = 11.84RR111 pKa = 11.84CYY113 pKa = 10.72SPNFFNSHH121 pKa = 4.16QAKK124 pKa = 10.27IPGEE128 pKa = 4.28VPASCDD134 pKa = 3.26EE135 pKa = 4.02NRR137 pKa = 11.84RR138 pKa = 11.84ASPFFLFARR147 pKa = 11.84IGG149 pKa = 3.34
Molecular weight: 16.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1385306 |
29 |
3557 |
347.5 |
39.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.85 ± 0.038 | 0.726 ± 0.013 |
5.561 ± 0.033 | 6.364 ± 0.045 |
4.954 ± 0.03 | 6.966 ± 0.033 |
2.032 ± 0.018 | 6.849 ± 0.033 |
6.742 ± 0.039 | 9.348 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.626 ± 0.018 | 5.179 ± 0.033 |
4.001 ± 0.022 | 3.553 ± 0.024 |
4.517 ± 0.033 | 6.218 ± 0.03 |
5.457 ± 0.036 | 6.721 ± 0.028 |
1.283 ± 0.018 | 4.053 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
