
Golden silk orbweaver associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Circularisvirus
Average proteome isoelectric point is 7.88
Get precalculated fractions of proteins
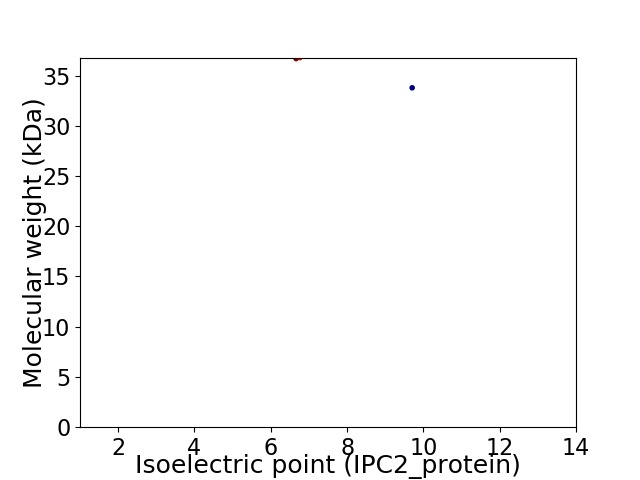
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPA5|A0A346BPA5_9VIRU Putative capsid protein OS=Golden silk orbweaver associated circular virus 1 OX=2293292 PE=4 SV=1
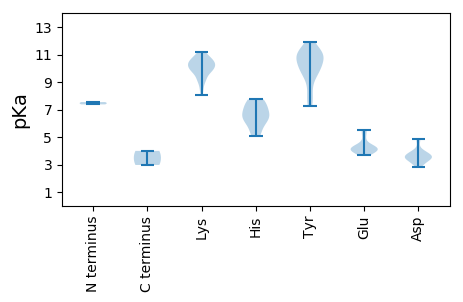
MM1 pKa = 7.37SKK3 pKa = 10.36CKK5 pKa = 9.84RR6 pKa = 11.84WLFTSYY12 pKa = 11.15NVVSEE17 pKa = 4.04PFYY20 pKa = 10.89DD21 pKa = 4.05EE22 pKa = 5.54KK23 pKa = 10.75IHH25 pKa = 7.24EE26 pKa = 4.32YY27 pKa = 10.83LCFGRR32 pKa = 11.84EE33 pKa = 4.14TCPSTNRR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 5.06LQGYY46 pKa = 7.61VAFKK50 pKa = 11.08NRR52 pKa = 11.84ITLAGIKK59 pKa = 9.56KK60 pKa = 8.59WIPGAHH66 pKa = 6.24FEE68 pKa = 4.31RR69 pKa = 11.84ARR71 pKa = 11.84GTPTEE76 pKa = 3.88NRR78 pKa = 11.84DD79 pKa = 3.38YY80 pKa = 10.97CKK82 pKa = 10.5KK83 pKa = 10.85DD84 pKa = 2.99GDD86 pKa = 3.91FKK88 pKa = 11.21EE89 pKa = 4.4WGSVPTVTGRR99 pKa = 11.84ACKK102 pKa = 10.35FGDD105 pKa = 3.43ILHH108 pKa = 6.64LAEE111 pKa = 5.39SGNITCIKK119 pKa = 9.24DD120 pKa = 3.54TYY122 pKa = 9.26PGVYY126 pKa = 9.82LRR128 pKa = 11.84YY129 pKa = 9.59KK130 pKa = 9.8ATLLSSLVCNTTDD143 pKa = 3.74LKK145 pKa = 11.13NSCGVWLCGPPRR157 pKa = 11.84SGKK160 pKa = 10.32DD161 pKa = 3.25YY162 pKa = 11.21AVRR165 pKa = 11.84QLDD168 pKa = 3.77NVYY171 pKa = 10.37MKK173 pKa = 10.06MLNKK177 pKa = 9.22WWDD180 pKa = 3.65GYY182 pKa = 11.21KK183 pKa = 10.49NEE185 pKa = 4.52DD186 pKa = 3.48NVLISDD192 pKa = 4.54IEE194 pKa = 4.17PDD196 pKa = 3.02HH197 pKa = 6.58GKK199 pKa = 9.66WIGYY203 pKa = 7.26YY204 pKa = 9.84IKK206 pKa = 9.91IWCDD210 pKa = 2.98MYY212 pKa = 11.66AFNAEE217 pKa = 4.01IKK219 pKa = 9.89GGSMKK224 pKa = 9.93IRR226 pKa = 11.84PKK228 pKa = 10.47KK229 pKa = 10.31IFMTSNYY236 pKa = 10.04KK237 pKa = 10.19LDD239 pKa = 3.62EE240 pKa = 4.36VFSGEE245 pKa = 3.97ILSAVAARR253 pKa = 11.84CNVYY257 pKa = 10.39DD258 pKa = 3.97YY259 pKa = 11.93SNGCDD264 pKa = 3.38VIVTKK269 pKa = 10.69RR270 pKa = 11.84MSPSPSDD277 pKa = 3.63VFISALKK284 pKa = 9.1EE285 pKa = 4.15HH286 pKa = 7.2EE287 pKa = 5.53DD288 pKa = 3.57GLSFTKK294 pKa = 10.3EE295 pKa = 3.82VLQTSSCLQEE305 pKa = 3.92EE306 pKa = 4.93AISSISEE313 pKa = 4.05EE314 pKa = 4.15SSNTCIEE321 pKa = 4.19EE322 pKa = 3.99
MM1 pKa = 7.37SKK3 pKa = 10.36CKK5 pKa = 9.84RR6 pKa = 11.84WLFTSYY12 pKa = 11.15NVVSEE17 pKa = 4.04PFYY20 pKa = 10.89DD21 pKa = 4.05EE22 pKa = 5.54KK23 pKa = 10.75IHH25 pKa = 7.24EE26 pKa = 4.32YY27 pKa = 10.83LCFGRR32 pKa = 11.84EE33 pKa = 4.14TCPSTNRR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 5.06LQGYY46 pKa = 7.61VAFKK50 pKa = 11.08NRR52 pKa = 11.84ITLAGIKK59 pKa = 9.56KK60 pKa = 8.59WIPGAHH66 pKa = 6.24FEE68 pKa = 4.31RR69 pKa = 11.84ARR71 pKa = 11.84GTPTEE76 pKa = 3.88NRR78 pKa = 11.84DD79 pKa = 3.38YY80 pKa = 10.97CKK82 pKa = 10.5KK83 pKa = 10.85DD84 pKa = 2.99GDD86 pKa = 3.91FKK88 pKa = 11.21EE89 pKa = 4.4WGSVPTVTGRR99 pKa = 11.84ACKK102 pKa = 10.35FGDD105 pKa = 3.43ILHH108 pKa = 6.64LAEE111 pKa = 5.39SGNITCIKK119 pKa = 9.24DD120 pKa = 3.54TYY122 pKa = 9.26PGVYY126 pKa = 9.82LRR128 pKa = 11.84YY129 pKa = 9.59KK130 pKa = 9.8ATLLSSLVCNTTDD143 pKa = 3.74LKK145 pKa = 11.13NSCGVWLCGPPRR157 pKa = 11.84SGKK160 pKa = 10.32DD161 pKa = 3.25YY162 pKa = 11.21AVRR165 pKa = 11.84QLDD168 pKa = 3.77NVYY171 pKa = 10.37MKK173 pKa = 10.06MLNKK177 pKa = 9.22WWDD180 pKa = 3.65GYY182 pKa = 11.21KK183 pKa = 10.49NEE185 pKa = 4.52DD186 pKa = 3.48NVLISDD192 pKa = 4.54IEE194 pKa = 4.17PDD196 pKa = 3.02HH197 pKa = 6.58GKK199 pKa = 9.66WIGYY203 pKa = 7.26YY204 pKa = 9.84IKK206 pKa = 9.91IWCDD210 pKa = 2.98MYY212 pKa = 11.66AFNAEE217 pKa = 4.01IKK219 pKa = 9.89GGSMKK224 pKa = 9.93IRR226 pKa = 11.84PKK228 pKa = 10.47KK229 pKa = 10.31IFMTSNYY236 pKa = 10.04KK237 pKa = 10.19LDD239 pKa = 3.62EE240 pKa = 4.36VFSGEE245 pKa = 3.97ILSAVAARR253 pKa = 11.84CNVYY257 pKa = 10.39DD258 pKa = 3.97YY259 pKa = 11.93SNGCDD264 pKa = 3.38VIVTKK269 pKa = 10.69RR270 pKa = 11.84MSPSPSDD277 pKa = 3.63VFISALKK284 pKa = 9.1EE285 pKa = 4.15HH286 pKa = 7.2EE287 pKa = 5.53DD288 pKa = 3.57GLSFTKK294 pKa = 10.3EE295 pKa = 3.82VLQTSSCLQEE305 pKa = 3.92EE306 pKa = 4.93AISSISEE313 pKa = 4.05EE314 pKa = 4.15SSNTCIEE321 pKa = 4.19EE322 pKa = 3.99
Molecular weight: 36.68 kDa
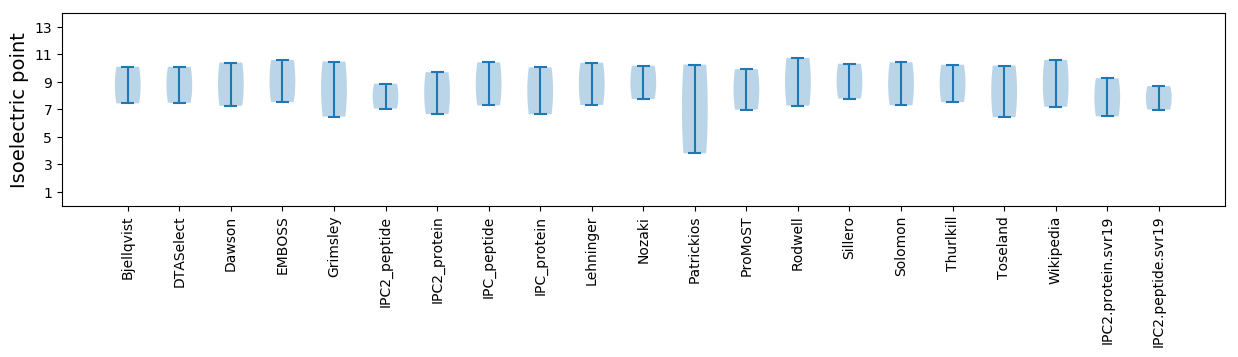
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPA5|A0A346BPA5_9VIRU Putative capsid protein OS=Golden silk orbweaver associated circular virus 1 OX=2293292 PE=4 SV=1
MM1 pKa = 7.54VYY3 pKa = 10.09RR4 pKa = 11.84SRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FYY10 pKa = 10.94KK11 pKa = 10.21RR12 pKa = 11.84RR13 pKa = 11.84LVYY16 pKa = 10.2KK17 pKa = 9.34KK18 pKa = 9.1RR19 pKa = 11.84QYY21 pKa = 10.78RR22 pKa = 11.84RR23 pKa = 11.84FRR25 pKa = 11.84RR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84IRR31 pKa = 11.84VLKK34 pKa = 10.39NKK36 pKa = 10.16RR37 pKa = 11.84SGNIYY42 pKa = 10.25KK43 pKa = 9.02LTVKK47 pKa = 10.48KK48 pKa = 10.82SFNTTLRR55 pKa = 11.84FYY57 pKa = 11.4VDD59 pKa = 3.25NASQSSVNITKK70 pKa = 9.94FEE72 pKa = 4.26RR73 pKa = 11.84DD74 pKa = 2.82IQMADD79 pKa = 3.28FFTNNHH85 pKa = 6.22EE86 pKa = 3.82IWDD89 pKa = 4.85RR90 pKa = 11.84IHH92 pKa = 7.54DD93 pKa = 3.57YY94 pKa = 11.01HH95 pKa = 7.73YY96 pKa = 10.99IKK98 pKa = 10.69FNYY101 pKa = 9.63FIIKK105 pKa = 9.83FPEE108 pKa = 3.69ITYY111 pKa = 8.23VTYY114 pKa = 10.89SGVTPTGIAKK124 pKa = 9.9QAVTGISSFNTEE136 pKa = 4.07RR137 pKa = 11.84YY138 pKa = 7.61PFHH141 pKa = 6.42VAWDD145 pKa = 3.78VEE147 pKa = 4.21QALDD151 pKa = 4.89FNAQDD156 pKa = 3.29ASFDD160 pKa = 3.65AKK162 pKa = 10.53EE163 pKa = 3.74LSEE166 pKa = 4.31YY167 pKa = 10.41PFAKK171 pKa = 9.74KK172 pKa = 10.26VYY174 pKa = 8.54PSQKK178 pKa = 8.43RR179 pKa = 11.84APYY182 pKa = 9.87FMWRR186 pKa = 11.84VPSCWRR192 pKa = 11.84QYY194 pKa = 10.89IDD196 pKa = 3.5TNAVQPLMFNNIPFYY211 pKa = 11.4NFFQNVTGIKK221 pKa = 9.85NVRR224 pKa = 11.84SPTKK228 pKa = 10.15ILGGHH233 pKa = 5.5EE234 pKa = 3.99NWWGNNLPSTVRR246 pKa = 11.84PDD248 pKa = 3.43IKK250 pKa = 10.68PSDD253 pKa = 3.78SDD255 pKa = 3.97TVFMQSFLKK264 pKa = 10.85CEE266 pKa = 5.09LIASVTFRR274 pKa = 11.84GVKK277 pKa = 8.06YY278 pKa = 9.33TGRR281 pKa = 11.84NSS283 pKa = 2.97
MM1 pKa = 7.54VYY3 pKa = 10.09RR4 pKa = 11.84SRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FYY10 pKa = 10.94KK11 pKa = 10.21RR12 pKa = 11.84RR13 pKa = 11.84LVYY16 pKa = 10.2KK17 pKa = 9.34KK18 pKa = 9.1RR19 pKa = 11.84QYY21 pKa = 10.78RR22 pKa = 11.84RR23 pKa = 11.84FRR25 pKa = 11.84RR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84IRR31 pKa = 11.84VLKK34 pKa = 10.39NKK36 pKa = 10.16RR37 pKa = 11.84SGNIYY42 pKa = 10.25KK43 pKa = 9.02LTVKK47 pKa = 10.48KK48 pKa = 10.82SFNTTLRR55 pKa = 11.84FYY57 pKa = 11.4VDD59 pKa = 3.25NASQSSVNITKK70 pKa = 9.94FEE72 pKa = 4.26RR73 pKa = 11.84DD74 pKa = 2.82IQMADD79 pKa = 3.28FFTNNHH85 pKa = 6.22EE86 pKa = 3.82IWDD89 pKa = 4.85RR90 pKa = 11.84IHH92 pKa = 7.54DD93 pKa = 3.57YY94 pKa = 11.01HH95 pKa = 7.73YY96 pKa = 10.99IKK98 pKa = 10.69FNYY101 pKa = 9.63FIIKK105 pKa = 9.83FPEE108 pKa = 3.69ITYY111 pKa = 8.23VTYY114 pKa = 10.89SGVTPTGIAKK124 pKa = 9.9QAVTGISSFNTEE136 pKa = 4.07RR137 pKa = 11.84YY138 pKa = 7.61PFHH141 pKa = 6.42VAWDD145 pKa = 3.78VEE147 pKa = 4.21QALDD151 pKa = 4.89FNAQDD156 pKa = 3.29ASFDD160 pKa = 3.65AKK162 pKa = 10.53EE163 pKa = 3.74LSEE166 pKa = 4.31YY167 pKa = 10.41PFAKK171 pKa = 9.74KK172 pKa = 10.26VYY174 pKa = 8.54PSQKK178 pKa = 8.43RR179 pKa = 11.84APYY182 pKa = 9.87FMWRR186 pKa = 11.84VPSCWRR192 pKa = 11.84QYY194 pKa = 10.89IDD196 pKa = 3.5TNAVQPLMFNNIPFYY211 pKa = 11.4NFFQNVTGIKK221 pKa = 9.85NVRR224 pKa = 11.84SPTKK228 pKa = 10.15ILGGHH233 pKa = 5.5EE234 pKa = 3.99NWWGNNLPSTVRR246 pKa = 11.84PDD248 pKa = 3.43IKK250 pKa = 10.68PSDD253 pKa = 3.78SDD255 pKa = 3.97TVFMQSFLKK264 pKa = 10.85CEE266 pKa = 5.09LIASVTFRR274 pKa = 11.84GVKK277 pKa = 8.06YY278 pKa = 9.33TGRR281 pKa = 11.84NSS283 pKa = 2.97
Molecular weight: 33.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
283 |
322 |
302.5 |
35.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.628 ± 0.022 | 2.645 ± 1.224 |
5.289 ± 0.439 | 5.124 ± 1.228 |
5.785 ± 1.48 | 5.289 ± 1.109 |
1.818 ± 0.032 | 6.446 ± 0.054 |
7.934 ± 0.324 | 5.289 ± 0.886 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.983 ± 0.137 | 5.95 ± 0.706 |
4.132 ± 0.292 | 2.479 ± 0.889 |
6.777 ± 1.523 | 8.099 ± 0.429 |
5.95 ± 0.259 | 6.281 ± 0.497 |
2.314 ± 0.123 | 5.785 ± 0.363 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
