
Bosea sp. LC85
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Boseaceae; Bosea; unclassified Bosea
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
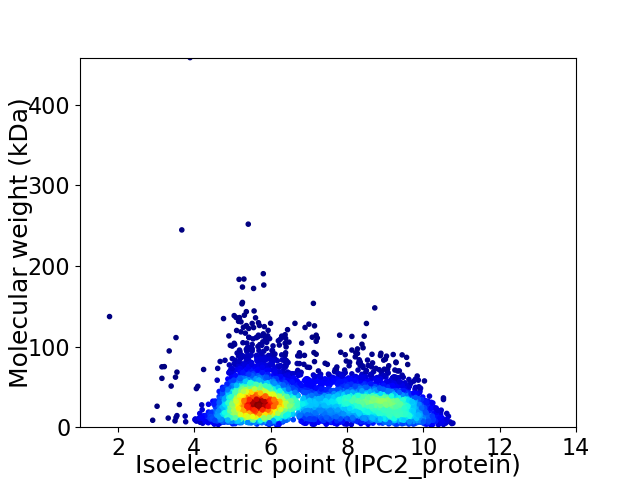
Virtual 2D-PAGE plot for 6190 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A085EUM5|A0A085EUM5_9BRAD Lipid A biosynthesis acyltransferase OS=Bosea sp. LC85 OX=1502851 GN=FG93_06022 PE=4 SV=1
MM1 pKa = 6.67TLSYY5 pKa = 10.22EE6 pKa = 4.21VSDD9 pKa = 3.85DD10 pKa = 3.72TVAVAQTAVFSVVAFTEE27 pKa = 3.94IDD29 pKa = 3.35GTASDD34 pKa = 4.93DD35 pKa = 3.85SLTGTEE41 pKa = 4.54CADD44 pKa = 5.07LIDD47 pKa = 4.92GRR49 pKa = 11.84DD50 pKa = 3.78GNDD53 pKa = 3.53TIDD56 pKa = 3.36GRR58 pKa = 11.84GGSDD62 pKa = 2.96ILHH65 pKa = 6.57GGAGNDD71 pKa = 4.64FIRR74 pKa = 11.84GGTGHH79 pKa = 7.32DD80 pKa = 4.42LIYY83 pKa = 10.67AGRR86 pKa = 11.84GNDD89 pKa = 2.86IVYY92 pKa = 10.54GGTGNDD98 pKa = 3.97TIHH101 pKa = 6.87GGEE104 pKa = 4.86GDD106 pKa = 3.78DD107 pKa = 4.53QLFGEE112 pKa = 5.34DD113 pKa = 3.76GDD115 pKa = 5.31DD116 pKa = 4.48IIFGGDD122 pKa = 3.38GNDD125 pKa = 3.93LLVGGNGNDD134 pKa = 3.63QLSGGAGNDD143 pKa = 3.72VIQGGAGDD151 pKa = 4.12DD152 pKa = 4.14QIAGDD157 pKa = 3.93AGDD160 pKa = 4.59DD161 pKa = 3.79VVHH164 pKa = 6.86GDD166 pKa = 4.17EE167 pKa = 6.15GDD169 pKa = 3.61DD170 pKa = 3.72TLDD173 pKa = 3.61GGVGNDD179 pKa = 4.16EE180 pKa = 4.26LHH182 pKa = 6.97GDD184 pKa = 3.65AGDD187 pKa = 3.67DD188 pKa = 3.52HH189 pKa = 8.7VMGGAGDD196 pKa = 3.81DD197 pKa = 3.88MIYY200 pKa = 10.87GGDD203 pKa = 3.8GDD205 pKa = 4.02DD206 pKa = 4.75TIDD209 pKa = 4.05GGADD213 pKa = 3.27SDD215 pKa = 4.74EE216 pKa = 4.5IDD218 pKa = 4.48AGEE221 pKa = 4.22GDD223 pKa = 3.59NTIFTGDD230 pKa = 3.39GNNSVRR236 pKa = 11.84TGNGSNLVTGGAGMDD251 pKa = 3.39HH252 pKa = 6.15VQFGSDD258 pKa = 2.78QDD260 pKa = 4.0TARR263 pKa = 11.84LGAGDD268 pKa = 4.49DD269 pKa = 3.76VAAGGGGNDD278 pKa = 3.55RR279 pKa = 11.84LFGEE283 pKa = 5.05AGDD286 pKa = 4.2DD287 pKa = 3.9TLSGEE292 pKa = 4.51AGDD295 pKa = 5.1DD296 pKa = 3.81VLDD299 pKa = 4.7GGAGTDD305 pKa = 3.72GVEE308 pKa = 4.56AGSGDD313 pKa = 3.65DD314 pKa = 4.49RR315 pKa = 11.84IVGTLDD321 pKa = 3.25AAADD325 pKa = 4.16CYY327 pKa = 10.66MGGSGDD333 pKa = 3.74DD334 pKa = 3.52TLDD337 pKa = 3.38YY338 pKa = 11.29SQATQSIDD346 pKa = 3.08INLVEE351 pKa = 4.53GKK353 pKa = 10.35AVGVEE358 pKa = 3.64IGEE361 pKa = 4.89DD362 pKa = 3.45GFSSIEE368 pKa = 3.93AVLGGSGDD376 pKa = 3.62DD377 pKa = 3.87HH378 pKa = 6.01FTIGAEE384 pKa = 3.89AVTLSGGAGGDD395 pKa = 3.48CFSFDD400 pKa = 3.51VPYY403 pKa = 11.02DD404 pKa = 3.68ADD406 pKa = 4.16DD407 pKa = 4.74RR408 pKa = 11.84DD409 pKa = 4.86LIHH412 pKa = 7.28QILDD416 pKa = 3.52LEE418 pKa = 5.14AGDD421 pKa = 4.51RR422 pKa = 11.84IVIKK426 pKa = 10.13QYY428 pKa = 9.8QIRR431 pKa = 11.84TDD433 pKa = 3.97DD434 pKa = 4.68DD435 pKa = 3.84PTGDD439 pKa = 3.73AAGDD443 pKa = 3.79GADD446 pKa = 4.58PFASTYY452 pKa = 10.98GDD454 pKa = 3.41EE455 pKa = 4.9ADD457 pKa = 4.4AAGQPFRR464 pKa = 11.84FRR466 pKa = 11.84IEE468 pKa = 4.21KK469 pKa = 10.12IGEE472 pKa = 3.98DD473 pKa = 3.86DD474 pKa = 3.49YY475 pKa = 11.64TYY477 pKa = 11.4VDD479 pKa = 3.9VYY481 pKa = 11.4LEE483 pKa = 4.0QTDD486 pKa = 3.67EE487 pKa = 4.14KK488 pKa = 10.68DD489 pKa = 3.48YY490 pKa = 11.21SIEE493 pKa = 3.84IQGSHH498 pKa = 6.15RR499 pKa = 11.84LYY501 pKa = 11.01YY502 pKa = 9.82YY503 pKa = 11.18
MM1 pKa = 6.67TLSYY5 pKa = 10.22EE6 pKa = 4.21VSDD9 pKa = 3.85DD10 pKa = 3.72TVAVAQTAVFSVVAFTEE27 pKa = 3.94IDD29 pKa = 3.35GTASDD34 pKa = 4.93DD35 pKa = 3.85SLTGTEE41 pKa = 4.54CADD44 pKa = 5.07LIDD47 pKa = 4.92GRR49 pKa = 11.84DD50 pKa = 3.78GNDD53 pKa = 3.53TIDD56 pKa = 3.36GRR58 pKa = 11.84GGSDD62 pKa = 2.96ILHH65 pKa = 6.57GGAGNDD71 pKa = 4.64FIRR74 pKa = 11.84GGTGHH79 pKa = 7.32DD80 pKa = 4.42LIYY83 pKa = 10.67AGRR86 pKa = 11.84GNDD89 pKa = 2.86IVYY92 pKa = 10.54GGTGNDD98 pKa = 3.97TIHH101 pKa = 6.87GGEE104 pKa = 4.86GDD106 pKa = 3.78DD107 pKa = 4.53QLFGEE112 pKa = 5.34DD113 pKa = 3.76GDD115 pKa = 5.31DD116 pKa = 4.48IIFGGDD122 pKa = 3.38GNDD125 pKa = 3.93LLVGGNGNDD134 pKa = 3.63QLSGGAGNDD143 pKa = 3.72VIQGGAGDD151 pKa = 4.12DD152 pKa = 4.14QIAGDD157 pKa = 3.93AGDD160 pKa = 4.59DD161 pKa = 3.79VVHH164 pKa = 6.86GDD166 pKa = 4.17EE167 pKa = 6.15GDD169 pKa = 3.61DD170 pKa = 3.72TLDD173 pKa = 3.61GGVGNDD179 pKa = 4.16EE180 pKa = 4.26LHH182 pKa = 6.97GDD184 pKa = 3.65AGDD187 pKa = 3.67DD188 pKa = 3.52HH189 pKa = 8.7VMGGAGDD196 pKa = 3.81DD197 pKa = 3.88MIYY200 pKa = 10.87GGDD203 pKa = 3.8GDD205 pKa = 4.02DD206 pKa = 4.75TIDD209 pKa = 4.05GGADD213 pKa = 3.27SDD215 pKa = 4.74EE216 pKa = 4.5IDD218 pKa = 4.48AGEE221 pKa = 4.22GDD223 pKa = 3.59NTIFTGDD230 pKa = 3.39GNNSVRR236 pKa = 11.84TGNGSNLVTGGAGMDD251 pKa = 3.39HH252 pKa = 6.15VQFGSDD258 pKa = 2.78QDD260 pKa = 4.0TARR263 pKa = 11.84LGAGDD268 pKa = 4.49DD269 pKa = 3.76VAAGGGGNDD278 pKa = 3.55RR279 pKa = 11.84LFGEE283 pKa = 5.05AGDD286 pKa = 4.2DD287 pKa = 3.9TLSGEE292 pKa = 4.51AGDD295 pKa = 5.1DD296 pKa = 3.81VLDD299 pKa = 4.7GGAGTDD305 pKa = 3.72GVEE308 pKa = 4.56AGSGDD313 pKa = 3.65DD314 pKa = 4.49RR315 pKa = 11.84IVGTLDD321 pKa = 3.25AAADD325 pKa = 4.16CYY327 pKa = 10.66MGGSGDD333 pKa = 3.74DD334 pKa = 3.52TLDD337 pKa = 3.38YY338 pKa = 11.29SQATQSIDD346 pKa = 3.08INLVEE351 pKa = 4.53GKK353 pKa = 10.35AVGVEE358 pKa = 3.64IGEE361 pKa = 4.89DD362 pKa = 3.45GFSSIEE368 pKa = 3.93AVLGGSGDD376 pKa = 3.62DD377 pKa = 3.87HH378 pKa = 6.01FTIGAEE384 pKa = 3.89AVTLSGGAGGDD395 pKa = 3.48CFSFDD400 pKa = 3.51VPYY403 pKa = 11.02DD404 pKa = 3.68ADD406 pKa = 4.16DD407 pKa = 4.74RR408 pKa = 11.84DD409 pKa = 4.86LIHH412 pKa = 7.28QILDD416 pKa = 3.52LEE418 pKa = 5.14AGDD421 pKa = 4.51RR422 pKa = 11.84IVIKK426 pKa = 10.13QYY428 pKa = 9.8QIRR431 pKa = 11.84TDD433 pKa = 3.97DD434 pKa = 4.68DD435 pKa = 3.84PTGDD439 pKa = 3.73AAGDD443 pKa = 3.79GADD446 pKa = 4.58PFASTYY452 pKa = 10.98GDD454 pKa = 3.41EE455 pKa = 4.9ADD457 pKa = 4.4AAGQPFRR464 pKa = 11.84FRR466 pKa = 11.84IEE468 pKa = 4.21KK469 pKa = 10.12IGEE472 pKa = 3.98DD473 pKa = 3.86DD474 pKa = 3.49YY475 pKa = 11.64TYY477 pKa = 11.4VDD479 pKa = 3.9VYY481 pKa = 11.4LEE483 pKa = 4.0QTDD486 pKa = 3.67EE487 pKa = 4.14KK488 pKa = 10.68DD489 pKa = 3.48YY490 pKa = 11.21SIEE493 pKa = 3.84IQGSHH498 pKa = 6.15RR499 pKa = 11.84LYY501 pKa = 11.01YY502 pKa = 9.82YY503 pKa = 11.18
Molecular weight: 51.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085FMT5|A0A085FMT5_9BRAD ABC transporter ATP-binding protein oligo/dipeptide transport protein OS=Bosea sp. LC85 OX=1502851 GN=FG93_02231 PE=3 SV=1
MM1 pKa = 7.45AADD4 pKa = 3.85KK5 pKa = 10.49PSRR8 pKa = 11.84APGRR12 pKa = 11.84LRR14 pKa = 11.84IVNPADD20 pKa = 3.19LTIRR24 pKa = 11.84RR25 pKa = 11.84VGRR28 pKa = 11.84GRR30 pKa = 11.84GFAYY34 pKa = 9.72IDD36 pKa = 3.9DD37 pKa = 4.48ADD39 pKa = 3.75ALVRR43 pKa = 11.84DD44 pKa = 3.87RR45 pKa = 11.84ATLEE49 pKa = 4.2RR50 pKa = 11.84IRR52 pKa = 11.84SLVIPPAYY60 pKa = 10.13CEE62 pKa = 3.95VRR64 pKa = 11.84IATDD68 pKa = 3.39PRR70 pKa = 11.84THH72 pKa = 6.25LQAIGRR78 pKa = 11.84DD79 pKa = 3.36EE80 pKa = 4.47AGRR83 pKa = 11.84LQYY86 pKa = 10.59LYY88 pKa = 9.89HH89 pKa = 6.94PEE91 pKa = 3.73WEE93 pKa = 4.19RR94 pKa = 11.84VRR96 pKa = 11.84EE97 pKa = 3.87RR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 10.1LKK102 pKa = 10.09RR103 pKa = 11.84LRR105 pKa = 11.84RR106 pKa = 11.84LIEE109 pKa = 3.8ALPRR113 pKa = 11.84LRR115 pKa = 11.84RR116 pKa = 11.84AVADD120 pKa = 3.94DD121 pKa = 4.1LKK123 pKa = 11.24ARR125 pKa = 11.84PLCRR129 pKa = 11.84RR130 pKa = 11.84KK131 pKa = 10.13ALACAVAIIDD141 pKa = 3.89RR142 pKa = 11.84CHH144 pKa = 6.67LRR146 pKa = 11.84IGGEE150 pKa = 4.16PYY152 pKa = 10.6AKK154 pKa = 10.56SNGSHH159 pKa = 6.47GASTLLKK166 pKa = 10.03RR167 pKa = 11.84HH168 pKa = 5.69VEE170 pKa = 4.11LTASRR175 pKa = 11.84IDD177 pKa = 3.34LLFRR181 pKa = 11.84GKK183 pKa = 10.28GGKK186 pKa = 9.24QIACSLQDD194 pKa = 3.32GSLARR199 pKa = 11.84ALGRR203 pKa = 11.84IAALPGRR210 pKa = 11.84RR211 pKa = 11.84LLQYY215 pKa = 10.31RR216 pKa = 11.84DD217 pKa = 3.65DD218 pKa = 5.37DD219 pKa = 4.17GTVVPVHH226 pKa = 6.35AADD229 pKa = 3.04INAYY233 pKa = 9.43LRR235 pKa = 11.84EE236 pKa = 4.03VSGLAISAKK245 pKa = 9.14DD246 pKa = 3.54LRR248 pKa = 11.84MLAGNAAAAEE258 pKa = 4.32LLLASQIITGDD269 pKa = 3.48GSEE272 pKa = 3.98KK273 pKa = 10.25RR274 pKa = 11.84QLAEE278 pKa = 3.52VMRR281 pKa = 11.84AVSEE285 pKa = 4.12QLANTPAVTRR295 pKa = 11.84KK296 pKa = 9.97SYY298 pKa = 10.06VHH300 pKa = 6.91AIVVNAYY307 pKa = 10.69ANGKK311 pKa = 8.82LAASFEE317 pKa = 4.37KK318 pKa = 10.6VRR320 pKa = 11.84AGGGCSRR327 pKa = 11.84IEE329 pKa = 3.68RR330 pKa = 11.84ALALLAAA337 pKa = 5.3
MM1 pKa = 7.45AADD4 pKa = 3.85KK5 pKa = 10.49PSRR8 pKa = 11.84APGRR12 pKa = 11.84LRR14 pKa = 11.84IVNPADD20 pKa = 3.19LTIRR24 pKa = 11.84RR25 pKa = 11.84VGRR28 pKa = 11.84GRR30 pKa = 11.84GFAYY34 pKa = 9.72IDD36 pKa = 3.9DD37 pKa = 4.48ADD39 pKa = 3.75ALVRR43 pKa = 11.84DD44 pKa = 3.87RR45 pKa = 11.84ATLEE49 pKa = 4.2RR50 pKa = 11.84IRR52 pKa = 11.84SLVIPPAYY60 pKa = 10.13CEE62 pKa = 3.95VRR64 pKa = 11.84IATDD68 pKa = 3.39PRR70 pKa = 11.84THH72 pKa = 6.25LQAIGRR78 pKa = 11.84DD79 pKa = 3.36EE80 pKa = 4.47AGRR83 pKa = 11.84LQYY86 pKa = 10.59LYY88 pKa = 9.89HH89 pKa = 6.94PEE91 pKa = 3.73WEE93 pKa = 4.19RR94 pKa = 11.84VRR96 pKa = 11.84EE97 pKa = 3.87RR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 10.1LKK102 pKa = 10.09RR103 pKa = 11.84LRR105 pKa = 11.84RR106 pKa = 11.84LIEE109 pKa = 3.8ALPRR113 pKa = 11.84LRR115 pKa = 11.84RR116 pKa = 11.84AVADD120 pKa = 3.94DD121 pKa = 4.1LKK123 pKa = 11.24ARR125 pKa = 11.84PLCRR129 pKa = 11.84RR130 pKa = 11.84KK131 pKa = 10.13ALACAVAIIDD141 pKa = 3.89RR142 pKa = 11.84CHH144 pKa = 6.67LRR146 pKa = 11.84IGGEE150 pKa = 4.16PYY152 pKa = 10.6AKK154 pKa = 10.56SNGSHH159 pKa = 6.47GASTLLKK166 pKa = 10.03RR167 pKa = 11.84HH168 pKa = 5.69VEE170 pKa = 4.11LTASRR175 pKa = 11.84IDD177 pKa = 3.34LLFRR181 pKa = 11.84GKK183 pKa = 10.28GGKK186 pKa = 9.24QIACSLQDD194 pKa = 3.32GSLARR199 pKa = 11.84ALGRR203 pKa = 11.84IAALPGRR210 pKa = 11.84RR211 pKa = 11.84LLQYY215 pKa = 10.31RR216 pKa = 11.84DD217 pKa = 3.65DD218 pKa = 5.37DD219 pKa = 4.17GTVVPVHH226 pKa = 6.35AADD229 pKa = 3.04INAYY233 pKa = 9.43LRR235 pKa = 11.84EE236 pKa = 4.03VSGLAISAKK245 pKa = 9.14DD246 pKa = 3.54LRR248 pKa = 11.84MLAGNAAAAEE258 pKa = 4.32LLLASQIITGDD269 pKa = 3.48GSEE272 pKa = 3.98KK273 pKa = 10.25RR274 pKa = 11.84QLAEE278 pKa = 3.52VMRR281 pKa = 11.84AVSEE285 pKa = 4.12QLANTPAVTRR295 pKa = 11.84KK296 pKa = 9.97SYY298 pKa = 10.06VHH300 pKa = 6.91AIVVNAYY307 pKa = 10.69ANGKK311 pKa = 8.82LAASFEE317 pKa = 4.37KK318 pKa = 10.6VRR320 pKa = 11.84AGGGCSRR327 pKa = 11.84IEE329 pKa = 3.68RR330 pKa = 11.84ALALLAAA337 pKa = 5.3
Molecular weight: 36.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1918328 |
29 |
4390 |
309.9 |
33.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.137 ± 0.045 | 0.812 ± 0.011 |
5.288 ± 0.033 | 5.44 ± 0.03 |
3.728 ± 0.022 | 8.856 ± 0.04 |
1.952 ± 0.016 | 5.22 ± 0.02 |
3.07 ± 0.031 | 10.519 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.017 | 2.376 ± 0.021 |
5.362 ± 0.024 | 3.055 ± 0.018 |
7.323 ± 0.032 | 5.454 ± 0.02 |
5.214 ± 0.025 | 7.432 ± 0.027 |
1.315 ± 0.013 | 2.074 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
