
Drosophila obscura sigmavirus 10A
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Drosophila obscura sigmavirus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
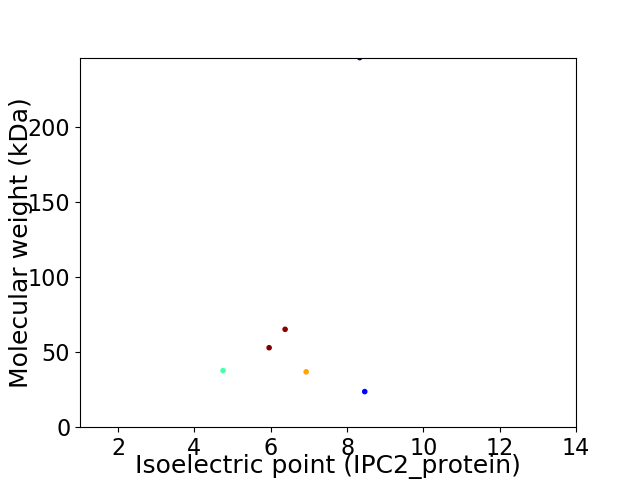
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8CJE8|C8CJE8_9RHAB Uncharacterized protein X OS=Drosophila obscura sigmavirus 10A OX=666962 GN=X PE=4 SV=1
MM1 pKa = 7.03STNPSKK7 pKa = 10.86PMNRR11 pKa = 11.84NSSEE15 pKa = 3.49KK16 pKa = 10.29DD17 pKa = 2.84IRR19 pKa = 11.84EE20 pKa = 4.0NSSSDD25 pKa = 3.26EE26 pKa = 4.08FEE28 pKa = 4.57EE29 pKa = 4.3EE30 pKa = 4.43VEE32 pKa = 4.23EE33 pKa = 4.38STSDD37 pKa = 3.49TEE39 pKa = 5.3KK40 pKa = 11.03NIPDD44 pKa = 5.43DD45 pKa = 3.75ISSPDD50 pKa = 3.57NEE52 pKa = 4.72FVPAPNQNIKK62 pKa = 10.59FSDD65 pKa = 3.42AFNSEE70 pKa = 4.13FGGKK74 pKa = 9.55YY75 pKa = 10.08NPEE78 pKa = 3.93EE79 pKa = 3.87LSKK82 pKa = 11.54YY83 pKa = 10.19GATDD87 pKa = 4.06DD88 pKa = 3.96YY89 pKa = 11.22CVAGRR94 pKa = 11.84LVQEE98 pKa = 4.24KK99 pKa = 10.93LNIKK103 pKa = 8.37EE104 pKa = 4.03TDD106 pKa = 3.18EE107 pKa = 5.25DD108 pKa = 3.84NVEE111 pKa = 4.0RR112 pKa = 11.84AILEE116 pKa = 4.13SSGVVPEE123 pKa = 5.08AEE125 pKa = 3.98AEE127 pKa = 4.06MEE129 pKa = 4.16EE130 pKa = 4.35LCEE133 pKa = 4.5KK134 pKa = 10.95SSDD137 pKa = 3.7EE138 pKa = 3.99NLEE141 pKa = 4.18CGATITLDD149 pKa = 4.12DD150 pKa = 5.08GFHH153 pKa = 6.36KK154 pKa = 10.31MKK156 pKa = 10.37IIASLEE162 pKa = 3.82EE163 pKa = 4.22RR164 pKa = 11.84FFDD167 pKa = 3.39PTQIHH172 pKa = 7.14LINRR176 pKa = 11.84FFRR179 pKa = 11.84EE180 pKa = 3.27LALYY184 pKa = 10.82VNTDD188 pKa = 3.26EE189 pKa = 4.63TEE191 pKa = 4.12GPILDD196 pKa = 3.53KK197 pKa = 11.12SKK199 pKa = 10.93NRR201 pKa = 11.84GAIVFGYY208 pKa = 10.37VEE210 pKa = 4.2SKK212 pKa = 9.8YY213 pKa = 10.54QKK215 pKa = 8.42KK216 pKa = 10.02QKK218 pKa = 7.61MTKK221 pKa = 10.18QNDD224 pKa = 3.5DD225 pKa = 4.68NIPSTSRR232 pKa = 11.84NCNKK236 pKa = 9.82NVCQPKK242 pKa = 9.89SPPPRR247 pKa = 11.84SEE249 pKa = 3.94GVAPAVNPNLQVILKK264 pKa = 9.76DD265 pKa = 3.73LNKK268 pKa = 9.97GLKK271 pKa = 8.03WKK273 pKa = 10.51HH274 pKa = 6.25FITQKK279 pKa = 10.35EE280 pKa = 4.3LSFKK284 pKa = 10.05MGQTGMTVEE293 pKa = 4.78EE294 pKa = 4.13ISAVLEE300 pKa = 4.23AGNEE304 pKa = 3.91IKK306 pKa = 10.52SGKK309 pKa = 10.17EE310 pKa = 3.33YY311 pKa = 11.15VEE313 pKa = 3.92YY314 pKa = 10.54LFKK317 pKa = 10.72IRR319 pKa = 11.84GHH321 pKa = 6.14FNQLEE326 pKa = 3.89ASYY329 pKa = 10.24IYY331 pKa = 10.51QPP333 pKa = 3.7
MM1 pKa = 7.03STNPSKK7 pKa = 10.86PMNRR11 pKa = 11.84NSSEE15 pKa = 3.49KK16 pKa = 10.29DD17 pKa = 2.84IRR19 pKa = 11.84EE20 pKa = 4.0NSSSDD25 pKa = 3.26EE26 pKa = 4.08FEE28 pKa = 4.57EE29 pKa = 4.3EE30 pKa = 4.43VEE32 pKa = 4.23EE33 pKa = 4.38STSDD37 pKa = 3.49TEE39 pKa = 5.3KK40 pKa = 11.03NIPDD44 pKa = 5.43DD45 pKa = 3.75ISSPDD50 pKa = 3.57NEE52 pKa = 4.72FVPAPNQNIKK62 pKa = 10.59FSDD65 pKa = 3.42AFNSEE70 pKa = 4.13FGGKK74 pKa = 9.55YY75 pKa = 10.08NPEE78 pKa = 3.93EE79 pKa = 3.87LSKK82 pKa = 11.54YY83 pKa = 10.19GATDD87 pKa = 4.06DD88 pKa = 3.96YY89 pKa = 11.22CVAGRR94 pKa = 11.84LVQEE98 pKa = 4.24KK99 pKa = 10.93LNIKK103 pKa = 8.37EE104 pKa = 4.03TDD106 pKa = 3.18EE107 pKa = 5.25DD108 pKa = 3.84NVEE111 pKa = 4.0RR112 pKa = 11.84AILEE116 pKa = 4.13SSGVVPEE123 pKa = 5.08AEE125 pKa = 3.98AEE127 pKa = 4.06MEE129 pKa = 4.16EE130 pKa = 4.35LCEE133 pKa = 4.5KK134 pKa = 10.95SSDD137 pKa = 3.7EE138 pKa = 3.99NLEE141 pKa = 4.18CGATITLDD149 pKa = 4.12DD150 pKa = 5.08GFHH153 pKa = 6.36KK154 pKa = 10.31MKK156 pKa = 10.37IIASLEE162 pKa = 3.82EE163 pKa = 4.22RR164 pKa = 11.84FFDD167 pKa = 3.39PTQIHH172 pKa = 7.14LINRR176 pKa = 11.84FFRR179 pKa = 11.84EE180 pKa = 3.27LALYY184 pKa = 10.82VNTDD188 pKa = 3.26EE189 pKa = 4.63TEE191 pKa = 4.12GPILDD196 pKa = 3.53KK197 pKa = 11.12SKK199 pKa = 10.93NRR201 pKa = 11.84GAIVFGYY208 pKa = 10.37VEE210 pKa = 4.2SKK212 pKa = 9.8YY213 pKa = 10.54QKK215 pKa = 8.42KK216 pKa = 10.02QKK218 pKa = 7.61MTKK221 pKa = 10.18QNDD224 pKa = 3.5DD225 pKa = 4.68NIPSTSRR232 pKa = 11.84NCNKK236 pKa = 9.82NVCQPKK242 pKa = 9.89SPPPRR247 pKa = 11.84SEE249 pKa = 3.94GVAPAVNPNLQVILKK264 pKa = 9.76DD265 pKa = 3.73LNKK268 pKa = 9.97GLKK271 pKa = 8.03WKK273 pKa = 10.51HH274 pKa = 6.25FITQKK279 pKa = 10.35EE280 pKa = 4.3LSFKK284 pKa = 10.05MGQTGMTVEE293 pKa = 4.78EE294 pKa = 4.13ISAVLEE300 pKa = 4.23AGNEE304 pKa = 3.91IKK306 pKa = 10.52SGKK309 pKa = 10.17EE310 pKa = 3.33YY311 pKa = 11.15VEE313 pKa = 3.92YY314 pKa = 10.54LFKK317 pKa = 10.72IRR319 pKa = 11.84GHH321 pKa = 6.14FNQLEE326 pKa = 3.89ASYY329 pKa = 10.24IYY331 pKa = 10.51QPP333 pKa = 3.7
Molecular weight: 37.67 kDa
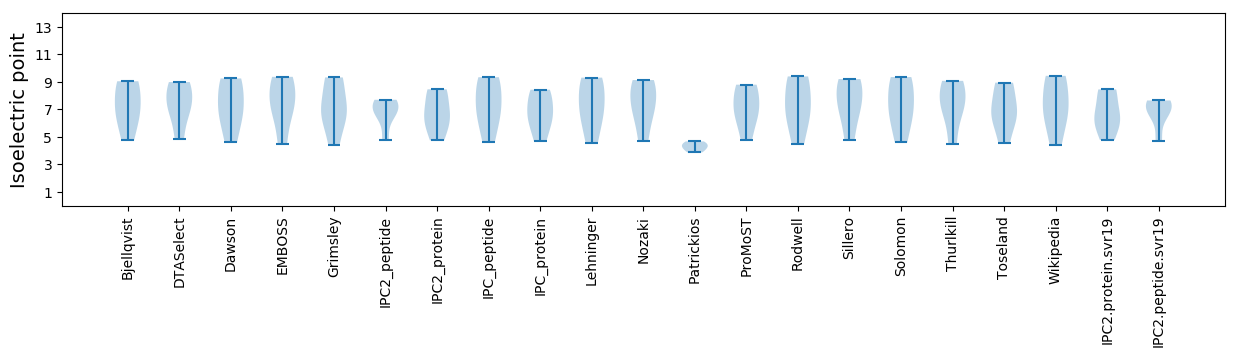
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8CJF0|C8CJF0_9RHAB Glycoprotein OS=Drosophila obscura sigmavirus 10A OX=666962 GN=G PE=4 SV=1
MM1 pKa = 7.37SKK3 pKa = 10.57KK4 pKa = 9.96EE5 pKa = 4.61AIPSLALMSTCIEE18 pKa = 4.09GKK20 pKa = 10.09EE21 pKa = 4.12EE22 pKa = 4.12IASSPHH28 pKa = 6.08SSFFVPTAPVNDD40 pKa = 4.47SIPYY44 pKa = 8.08YY45 pKa = 9.88VHH47 pKa = 6.69NLTVEE52 pKa = 4.13AEE54 pKa = 3.87ISVNSNRR61 pKa = 11.84VIDD64 pKa = 3.43TWEE67 pKa = 4.0PLRR70 pKa = 11.84EE71 pKa = 4.02ILTSWVDD78 pKa = 3.44NYY80 pKa = 11.02KK81 pKa = 11.0GPLNVKK87 pKa = 9.99SILGVVYY94 pKa = 9.62TLLIFKK100 pKa = 8.2MKK102 pKa = 10.11KK103 pKa = 8.3MRR105 pKa = 11.84VGVNYY110 pKa = 10.5LMYY113 pKa = 10.17GIKK116 pKa = 9.56MFDD119 pKa = 3.57RR120 pKa = 11.84ISFLMTYY127 pKa = 9.82NFEE130 pKa = 4.03NSIPVQGEE138 pKa = 3.94LNYY141 pKa = 10.7KK142 pKa = 9.66SGRR145 pKa = 11.84GQFLYY150 pKa = 10.13TVNFKK155 pKa = 10.93YY156 pKa = 10.47RR157 pKa = 11.84LTPIKK162 pKa = 10.34RR163 pKa = 11.84HH164 pKa = 5.37GSLIQTLFSPGVVDD178 pKa = 3.32MSKK181 pKa = 10.4QVGPTLAVALDD192 pKa = 3.93SIGIKK197 pKa = 9.36TKK199 pKa = 10.34EE200 pKa = 4.09DD201 pKa = 3.49PAGLVLLYY209 pKa = 7.51TTT211 pKa = 4.93
MM1 pKa = 7.37SKK3 pKa = 10.57KK4 pKa = 9.96EE5 pKa = 4.61AIPSLALMSTCIEE18 pKa = 4.09GKK20 pKa = 10.09EE21 pKa = 4.12EE22 pKa = 4.12IASSPHH28 pKa = 6.08SSFFVPTAPVNDD40 pKa = 4.47SIPYY44 pKa = 8.08YY45 pKa = 9.88VHH47 pKa = 6.69NLTVEE52 pKa = 4.13AEE54 pKa = 3.87ISVNSNRR61 pKa = 11.84VIDD64 pKa = 3.43TWEE67 pKa = 4.0PLRR70 pKa = 11.84EE71 pKa = 4.02ILTSWVDD78 pKa = 3.44NYY80 pKa = 11.02KK81 pKa = 11.0GPLNVKK87 pKa = 9.99SILGVVYY94 pKa = 9.62TLLIFKK100 pKa = 8.2MKK102 pKa = 10.11KK103 pKa = 8.3MRR105 pKa = 11.84VGVNYY110 pKa = 10.5LMYY113 pKa = 10.17GIKK116 pKa = 9.56MFDD119 pKa = 3.57RR120 pKa = 11.84ISFLMTYY127 pKa = 9.82NFEE130 pKa = 4.03NSIPVQGEE138 pKa = 3.94LNYY141 pKa = 10.7KK142 pKa = 9.66SGRR145 pKa = 11.84GQFLYY150 pKa = 10.13TVNFKK155 pKa = 10.93YY156 pKa = 10.47RR157 pKa = 11.84LTPIKK162 pKa = 10.34RR163 pKa = 11.84HH164 pKa = 5.37GSLIQTLFSPGVVDD178 pKa = 3.32MSKK181 pKa = 10.4QVGPTLAVALDD192 pKa = 3.93SIGIKK197 pKa = 9.36TKK199 pKa = 10.34EE200 pKa = 4.09DD201 pKa = 3.49PAGLVLLYY209 pKa = 7.51TTT211 pKa = 4.93
Molecular weight: 23.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4052 |
211 |
2146 |
675.3 |
77.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.763 ± 0.769 | 1.234 ± 0.159 |
5.01 ± 0.198 | 6.318 ± 0.881 |
4.22 ± 0.175 | 5.503 ± 0.353 |
2.468 ± 0.356 | 7.28 ± 0.481 |
6.885 ± 0.338 | 10.242 ± 0.692 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.295 ± 0.324 | 5.528 ± 0.378 |
4.541 ± 0.34 | 3.258 ± 0.297 |
4.837 ± 0.518 | 8.119 ± 0.365 |
6.145 ± 0.29 | 5.602 ± 0.483 |
1.9 ± 0.255 | 3.85 ± 0.318 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
