
Aspergillus spelaeus tetramycovirus 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus; Aspergillus spelaeus polymycovirus 1
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
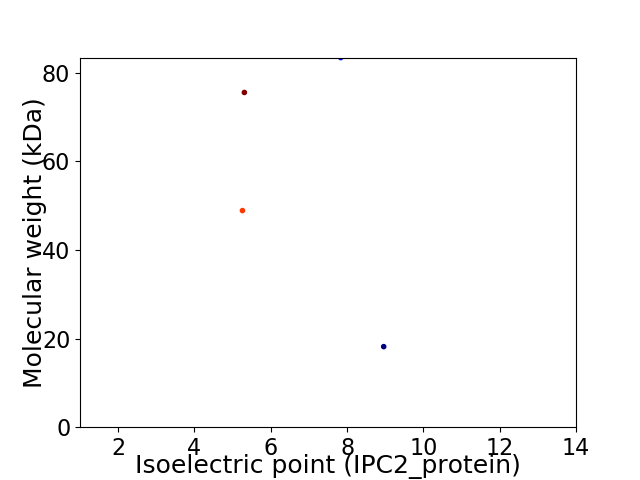
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G3C4N7|A0A3G3C4N7_9VIRU Uncharacterized protein OS=Aspergillus spelaeus tetramycovirus 1 OX=2485922 PE=4 SV=1
MM1 pKa = 7.84ADD3 pKa = 3.99LARR6 pKa = 11.84LRR8 pKa = 11.84AIALSGDD15 pKa = 3.42LTVNLTAAILHH26 pKa = 6.34HH27 pKa = 6.71LSPDD31 pKa = 3.14QSEE34 pKa = 4.27TTRR37 pKa = 11.84YY38 pKa = 9.96GIADD42 pKa = 3.61VRR44 pKa = 11.84EE45 pKa = 4.02VMTWLSRR52 pKa = 11.84APGQFYY58 pKa = 11.02GDD60 pKa = 3.4VVAATGVTPAILSAIVDD77 pKa = 3.83EE78 pKa = 5.02MDD80 pKa = 4.89DD81 pKa = 3.9AACADD86 pKa = 3.99AATMANAMATPDD98 pKa = 3.89AEE100 pKa = 4.09WDD102 pKa = 3.55RR103 pKa = 11.84EE104 pKa = 3.95HH105 pKa = 7.58RR106 pKa = 11.84NRR108 pKa = 11.84FTTALATHH116 pKa = 6.39SADD119 pKa = 3.61AARR122 pKa = 11.84LAAQSSAEE130 pKa = 3.96FAALDD135 pKa = 3.73SLVEE139 pKa = 3.93AGYY142 pKa = 11.22ANEE145 pKa = 4.0DD146 pKa = 3.52HH147 pKa = 7.09ARR149 pKa = 11.84TAVNEE154 pKa = 3.91MNRR157 pKa = 11.84AYY159 pKa = 10.22GVNYY163 pKa = 10.02RR164 pKa = 11.84VLTRR168 pKa = 11.84QRR170 pKa = 11.84ARR172 pKa = 11.84SCVAIAVKK180 pKa = 10.5YY181 pKa = 9.9RR182 pKa = 11.84LPDD185 pKa = 3.22LTTAHH190 pKa = 7.48ILTPWCLSRR199 pKa = 11.84TAAMAYY205 pKa = 7.81VAVVARR211 pKa = 11.84HH212 pKa = 5.36IRR214 pKa = 11.84RR215 pKa = 11.84QQSEE219 pKa = 3.7ARR221 pKa = 11.84AGRR224 pKa = 11.84IARR227 pKa = 11.84TALSYY232 pKa = 11.09ASRR235 pKa = 11.84SGLDD239 pKa = 3.08AVRR242 pKa = 11.84SVVEE246 pKa = 3.76PAVAILTSFRR256 pKa = 11.84YY257 pKa = 9.66DD258 pKa = 3.2AEE260 pKa = 3.81RR261 pKa = 11.84LAFVDD266 pKa = 3.24TFDD269 pKa = 5.15RR270 pKa = 11.84EE271 pKa = 4.3RR272 pKa = 11.84EE273 pKa = 4.34SITTRR278 pKa = 11.84ASSVLVAFACIHH290 pKa = 5.64MCGEE294 pKa = 4.02TDD296 pKa = 3.59VLIRR300 pKa = 11.84AAQLRR305 pKa = 11.84ADD307 pKa = 4.06VVPDD311 pKa = 3.17QTPPHH316 pKa = 6.7PSVVEE321 pKa = 3.9YY322 pKa = 10.86TEE324 pKa = 4.33TVDD327 pKa = 5.69DD328 pKa = 4.41EE329 pKa = 4.59QTGLLMFSRR338 pKa = 11.84LLHH341 pKa = 6.1AARR344 pKa = 11.84AQRR347 pKa = 11.84DD348 pKa = 3.44QSRR351 pKa = 11.84AGPRR355 pKa = 11.84VNKK358 pKa = 10.24DD359 pKa = 2.92LMLGYY364 pKa = 9.82VARR367 pKa = 11.84GKK369 pKa = 10.45KK370 pKa = 9.66KK371 pKa = 10.29SEE373 pKa = 4.28RR374 pKa = 11.84NTMACINRR382 pKa = 11.84VEE384 pKa = 4.25EE385 pKa = 4.21AVSLLRR391 pKa = 11.84SKK393 pKa = 10.78KK394 pKa = 10.36ADD396 pKa = 3.35VLNTLFVIEE405 pKa = 4.4WGGEE409 pKa = 3.63FDD411 pKa = 4.36YY412 pKa = 9.22PTVMAALAVARR423 pKa = 11.84LDD425 pKa = 3.48VCVDD429 pKa = 3.7IGPSGIDD436 pKa = 3.33LPGADD441 pKa = 4.06VVADD445 pKa = 4.15DD446 pKa = 4.23TPEE449 pKa = 3.97EE450 pKa = 4.22YY451 pKa = 10.38NYY453 pKa = 11.42SLLISSAEE461 pKa = 3.82EE462 pKa = 3.52RR463 pKa = 11.84MMPRR467 pKa = 11.84MPRR470 pKa = 11.84ISYY473 pKa = 9.75PKK475 pKa = 10.23DD476 pKa = 3.21EE477 pKa = 4.63PLEE480 pKa = 4.46SKK482 pKa = 10.53LLRR485 pKa = 11.84VAEE488 pKa = 4.84FYY490 pKa = 11.19CRR492 pKa = 11.84DD493 pKa = 3.21GRR495 pKa = 11.84SNIAYY500 pKa = 9.5ISGGTLQHH508 pKa = 6.63EE509 pKa = 4.75GTPVSVCVDD518 pKa = 3.23TNNRR522 pKa = 11.84LDD524 pKa = 4.52AIRR527 pKa = 11.84NATAQVPINFACAEE541 pKa = 4.01ALIPPLCPHH550 pKa = 7.3GAQAPLEE557 pKa = 4.46EE558 pKa = 4.32YY559 pKa = 10.86LDD561 pKa = 3.78TSSLIDD567 pKa = 4.05EE568 pKa = 4.84EE569 pKa = 5.0CPNCNAHH576 pKa = 5.71YY577 pKa = 10.29RR578 pKa = 11.84SVALLSAACAKK589 pKa = 10.33GHH591 pKa = 5.0TRR593 pKa = 11.84VVKK596 pKa = 10.03PRR598 pKa = 11.84SSYY601 pKa = 9.95AHH603 pKa = 6.33NFHH606 pKa = 6.79VSLEE610 pKa = 4.25LVPGHH615 pKa = 5.88PTCPEE620 pKa = 3.93DD621 pKa = 3.49TLTTIDD627 pKa = 4.03SMVAANVCRR636 pKa = 11.84NADD639 pKa = 3.3WGDD642 pKa = 3.65TPNFPLGGGNPSSEE656 pKa = 4.37NYY658 pKa = 9.63RR659 pKa = 11.84KK660 pKa = 10.41LNAITMKK667 pKa = 10.34HH668 pKa = 5.55VYY670 pKa = 10.27ACLSSVATGPFDD682 pKa = 4.41YY683 pKa = 11.22DD684 pKa = 3.78DD685 pKa = 4.1MASIAASIAPP695 pKa = 3.85
MM1 pKa = 7.84ADD3 pKa = 3.99LARR6 pKa = 11.84LRR8 pKa = 11.84AIALSGDD15 pKa = 3.42LTVNLTAAILHH26 pKa = 6.34HH27 pKa = 6.71LSPDD31 pKa = 3.14QSEE34 pKa = 4.27TTRR37 pKa = 11.84YY38 pKa = 9.96GIADD42 pKa = 3.61VRR44 pKa = 11.84EE45 pKa = 4.02VMTWLSRR52 pKa = 11.84APGQFYY58 pKa = 11.02GDD60 pKa = 3.4VVAATGVTPAILSAIVDD77 pKa = 3.83EE78 pKa = 5.02MDD80 pKa = 4.89DD81 pKa = 3.9AACADD86 pKa = 3.99AATMANAMATPDD98 pKa = 3.89AEE100 pKa = 4.09WDD102 pKa = 3.55RR103 pKa = 11.84EE104 pKa = 3.95HH105 pKa = 7.58RR106 pKa = 11.84NRR108 pKa = 11.84FTTALATHH116 pKa = 6.39SADD119 pKa = 3.61AARR122 pKa = 11.84LAAQSSAEE130 pKa = 3.96FAALDD135 pKa = 3.73SLVEE139 pKa = 3.93AGYY142 pKa = 11.22ANEE145 pKa = 4.0DD146 pKa = 3.52HH147 pKa = 7.09ARR149 pKa = 11.84TAVNEE154 pKa = 3.91MNRR157 pKa = 11.84AYY159 pKa = 10.22GVNYY163 pKa = 10.02RR164 pKa = 11.84VLTRR168 pKa = 11.84QRR170 pKa = 11.84ARR172 pKa = 11.84SCVAIAVKK180 pKa = 10.5YY181 pKa = 9.9RR182 pKa = 11.84LPDD185 pKa = 3.22LTTAHH190 pKa = 7.48ILTPWCLSRR199 pKa = 11.84TAAMAYY205 pKa = 7.81VAVVARR211 pKa = 11.84HH212 pKa = 5.36IRR214 pKa = 11.84RR215 pKa = 11.84QQSEE219 pKa = 3.7ARR221 pKa = 11.84AGRR224 pKa = 11.84IARR227 pKa = 11.84TALSYY232 pKa = 11.09ASRR235 pKa = 11.84SGLDD239 pKa = 3.08AVRR242 pKa = 11.84SVVEE246 pKa = 3.76PAVAILTSFRR256 pKa = 11.84YY257 pKa = 9.66DD258 pKa = 3.2AEE260 pKa = 3.81RR261 pKa = 11.84LAFVDD266 pKa = 3.24TFDD269 pKa = 5.15RR270 pKa = 11.84EE271 pKa = 4.3RR272 pKa = 11.84EE273 pKa = 4.34SITTRR278 pKa = 11.84ASSVLVAFACIHH290 pKa = 5.64MCGEE294 pKa = 4.02TDD296 pKa = 3.59VLIRR300 pKa = 11.84AAQLRR305 pKa = 11.84ADD307 pKa = 4.06VVPDD311 pKa = 3.17QTPPHH316 pKa = 6.7PSVVEE321 pKa = 3.9YY322 pKa = 10.86TEE324 pKa = 4.33TVDD327 pKa = 5.69DD328 pKa = 4.41EE329 pKa = 4.59QTGLLMFSRR338 pKa = 11.84LLHH341 pKa = 6.1AARR344 pKa = 11.84AQRR347 pKa = 11.84DD348 pKa = 3.44QSRR351 pKa = 11.84AGPRR355 pKa = 11.84VNKK358 pKa = 10.24DD359 pKa = 2.92LMLGYY364 pKa = 9.82VARR367 pKa = 11.84GKK369 pKa = 10.45KK370 pKa = 9.66KK371 pKa = 10.29SEE373 pKa = 4.28RR374 pKa = 11.84NTMACINRR382 pKa = 11.84VEE384 pKa = 4.25EE385 pKa = 4.21AVSLLRR391 pKa = 11.84SKK393 pKa = 10.78KK394 pKa = 10.36ADD396 pKa = 3.35VLNTLFVIEE405 pKa = 4.4WGGEE409 pKa = 3.63FDD411 pKa = 4.36YY412 pKa = 9.22PTVMAALAVARR423 pKa = 11.84LDD425 pKa = 3.48VCVDD429 pKa = 3.7IGPSGIDD436 pKa = 3.33LPGADD441 pKa = 4.06VVADD445 pKa = 4.15DD446 pKa = 4.23TPEE449 pKa = 3.97EE450 pKa = 4.22YY451 pKa = 10.38NYY453 pKa = 11.42SLLISSAEE461 pKa = 3.82EE462 pKa = 3.52RR463 pKa = 11.84MMPRR467 pKa = 11.84MPRR470 pKa = 11.84ISYY473 pKa = 9.75PKK475 pKa = 10.23DD476 pKa = 3.21EE477 pKa = 4.63PLEE480 pKa = 4.46SKK482 pKa = 10.53LLRR485 pKa = 11.84VAEE488 pKa = 4.84FYY490 pKa = 11.19CRR492 pKa = 11.84DD493 pKa = 3.21GRR495 pKa = 11.84SNIAYY500 pKa = 9.5ISGGTLQHH508 pKa = 6.63EE509 pKa = 4.75GTPVSVCVDD518 pKa = 3.23TNNRR522 pKa = 11.84LDD524 pKa = 4.52AIRR527 pKa = 11.84NATAQVPINFACAEE541 pKa = 4.01ALIPPLCPHH550 pKa = 7.3GAQAPLEE557 pKa = 4.46EE558 pKa = 4.32YY559 pKa = 10.86LDD561 pKa = 3.78TSSLIDD567 pKa = 4.05EE568 pKa = 4.84EE569 pKa = 5.0CPNCNAHH576 pKa = 5.71YY577 pKa = 10.29RR578 pKa = 11.84SVALLSAACAKK589 pKa = 10.33GHH591 pKa = 5.0TRR593 pKa = 11.84VVKK596 pKa = 10.03PRR598 pKa = 11.84SSYY601 pKa = 9.95AHH603 pKa = 6.33NFHH606 pKa = 6.79VSLEE610 pKa = 4.25LVPGHH615 pKa = 5.88PTCPEE620 pKa = 3.93DD621 pKa = 3.49TLTTIDD627 pKa = 4.03SMVAANVCRR636 pKa = 11.84NADD639 pKa = 3.3WGDD642 pKa = 3.65TPNFPLGGGNPSSEE656 pKa = 4.37NYY658 pKa = 9.63RR659 pKa = 11.84KK660 pKa = 10.41LNAITMKK667 pKa = 10.34HH668 pKa = 5.55VYY670 pKa = 10.27ACLSSVATGPFDD682 pKa = 4.41YY683 pKa = 11.22DD684 pKa = 3.78DD685 pKa = 4.1MASIAASIAPP695 pKa = 3.85
Molecular weight: 75.58 kDa
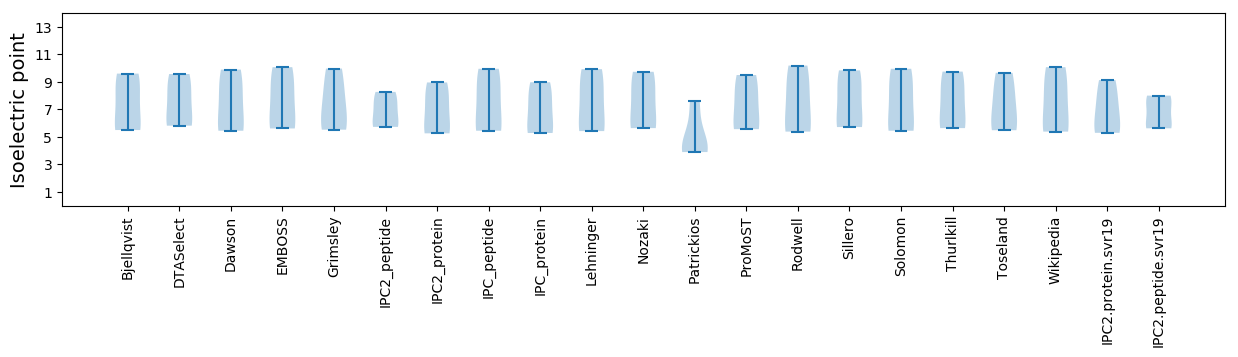
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G3C4N5|A0A3G3C4N5_9VIRU Methyltransferase OS=Aspergillus spelaeus tetramycovirus 1 OX=2485922 PE=4 SV=1
MM1 pKa = 8.04DD2 pKa = 6.43LLHH5 pKa = 7.2SDD7 pKa = 4.33PEE9 pKa = 4.36QACSRR14 pKa = 11.84IHH16 pKa = 6.42GLVSDD21 pKa = 4.7RR22 pKa = 11.84LRR24 pKa = 11.84KK25 pKa = 9.65RR26 pKa = 11.84GSSRR30 pKa = 11.84PVKK33 pKa = 10.43VDD35 pKa = 3.59LEE37 pKa = 4.39GSPSAKK43 pKa = 10.03SAGNSAPSSGDD54 pKa = 2.8GGLALLRR61 pKa = 11.84RR62 pKa = 11.84EE63 pKa = 4.41VKK65 pKa = 10.42RR66 pKa = 11.84NSGVYY71 pKa = 10.17GSYY74 pKa = 10.33QFAAGPTKK82 pKa = 10.61RR83 pKa = 11.84PGGQAFSVDD92 pKa = 3.5LGGGLWALGQSKK104 pKa = 10.24AVAVAAARR112 pKa = 11.84ICRR115 pKa = 11.84VLGRR119 pKa = 11.84HH120 pKa = 5.89EE121 pKa = 4.86DD122 pKa = 3.37LVRR125 pKa = 11.84PYY127 pKa = 10.21IYY129 pKa = 10.29YY130 pKa = 8.69WASGSSPRR138 pKa = 11.84SAADD142 pKa = 2.85ISEE145 pKa = 4.35KK146 pKa = 10.67VWDD149 pKa = 4.18GRR151 pKa = 11.84LGPHH155 pKa = 6.61EE156 pKa = 5.03DD157 pKa = 4.25APPEE161 pKa = 4.15PATKK165 pKa = 10.49AKK167 pKa = 10.11TEE169 pKa = 3.93ATKK172 pKa = 10.91
MM1 pKa = 8.04DD2 pKa = 6.43LLHH5 pKa = 7.2SDD7 pKa = 4.33PEE9 pKa = 4.36QACSRR14 pKa = 11.84IHH16 pKa = 6.42GLVSDD21 pKa = 4.7RR22 pKa = 11.84LRR24 pKa = 11.84KK25 pKa = 9.65RR26 pKa = 11.84GSSRR30 pKa = 11.84PVKK33 pKa = 10.43VDD35 pKa = 3.59LEE37 pKa = 4.39GSPSAKK43 pKa = 10.03SAGNSAPSSGDD54 pKa = 2.8GGLALLRR61 pKa = 11.84RR62 pKa = 11.84EE63 pKa = 4.41VKK65 pKa = 10.42RR66 pKa = 11.84NSGVYY71 pKa = 10.17GSYY74 pKa = 10.33QFAAGPTKK82 pKa = 10.61RR83 pKa = 11.84PGGQAFSVDD92 pKa = 3.5LGGGLWALGQSKK104 pKa = 10.24AVAVAAARR112 pKa = 11.84ICRR115 pKa = 11.84VLGRR119 pKa = 11.84HH120 pKa = 5.89EE121 pKa = 4.86DD122 pKa = 3.37LVRR125 pKa = 11.84PYY127 pKa = 10.21IYY129 pKa = 10.29YY130 pKa = 8.69WASGSSPRR138 pKa = 11.84SAADD142 pKa = 2.85ISEE145 pKa = 4.35KK146 pKa = 10.67VWDD149 pKa = 4.18GRR151 pKa = 11.84LGPHH155 pKa = 6.61EE156 pKa = 5.03DD157 pKa = 4.25APPEE161 pKa = 4.15PATKK165 pKa = 10.49AKK167 pKa = 10.11TEE169 pKa = 3.93ATKK172 pKa = 10.91
Molecular weight: 18.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2075 |
172 |
760 |
518.8 |
56.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.422 ± 1.521 | 1.253 ± 0.491 |
7.181 ± 0.447 | 4.916 ± 0.356 |
2.843 ± 0.442 | 7.229 ± 1.304 |
2.602 ± 0.239 | 3.181 ± 0.507 |
3.229 ± 0.665 | 9.542 ± 0.688 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.41 ± 0.582 | 2.554 ± 0.592 |
5.928 ± 0.538 | 1.928 ± 0.101 |
7.855 ± 0.727 | 7.518 ± 0.924 |
5.446 ± 0.653 | 8.53 ± 1.172 |
0.867 ± 0.157 | 3.566 ± 0.233 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
