
Candidatus Mycoplasma haemobos
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
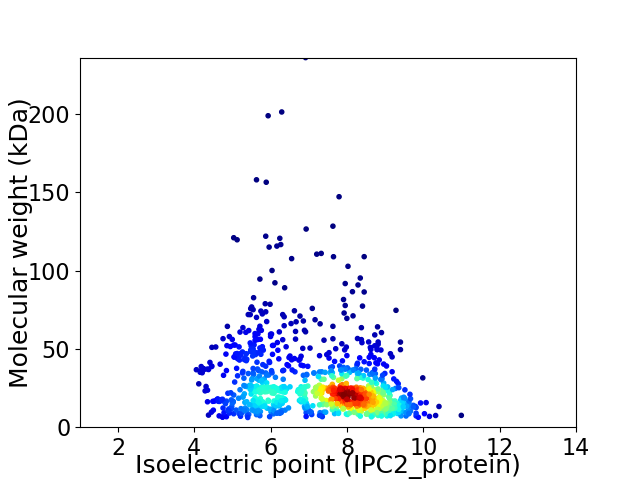
Virtual 2D-PAGE plot for 1109 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A9QDZ4|A0A1A9QDZ4_9MOLU Uncharacterized protein OS=Candidatus Mycoplasma haemobos OX=432608 GN=A6V39_00240 PE=4 SV=1
MM1 pKa = 7.4LPVATVTTATILTTSLISSGKK22 pKa = 9.89VDD24 pKa = 4.52LDD26 pKa = 3.44KK27 pKa = 11.4KK28 pKa = 10.95KK29 pKa = 10.83EE30 pKa = 4.04NEE32 pKa = 4.01DD33 pKa = 3.31QEE35 pKa = 5.8LNDD38 pKa = 3.84FVKK41 pKa = 10.63PPEE44 pKa = 4.34DD45 pKa = 4.09FEE47 pKa = 4.77DD48 pKa = 3.56EE49 pKa = 4.36KK50 pKa = 11.08IFYY53 pKa = 10.5SKK55 pKa = 11.03EE56 pKa = 3.75GDD58 pKa = 3.48SDD60 pKa = 3.84NLRR63 pKa = 11.84TLDD66 pKa = 3.83KK67 pKa = 10.54DD68 pKa = 3.6TAKK71 pKa = 10.7KK72 pKa = 10.15IEE74 pKa = 4.17EE75 pKa = 4.05LMQKK79 pKa = 10.38NGGKK83 pKa = 9.76LEE85 pKa = 4.28KK86 pKa = 10.8GEE88 pKa = 4.03QLGDD92 pKa = 3.46TVQAPNPEE100 pKa = 4.11INPNVPEE107 pKa = 4.1NQPQNVGKK115 pKa = 10.26ADD117 pKa = 3.68SPEE120 pKa = 3.97ITSKK124 pKa = 11.48AEE126 pKa = 4.24DD127 pKa = 3.61KK128 pKa = 11.49DD129 pKa = 3.67MTSVPKK135 pKa = 10.82GNNKK139 pKa = 10.3DD140 pKa = 3.5EE141 pKa = 4.83GDD143 pKa = 4.1TISEE147 pKa = 4.4TKK149 pKa = 9.54DD150 pKa = 3.17TLISTSEE157 pKa = 4.02STDD160 pKa = 3.51DD161 pKa = 4.67DD162 pKa = 4.08FTNGRR167 pKa = 11.84DD168 pKa = 4.0DD169 pKa = 4.01PQQPSLDD176 pKa = 4.04DD177 pKa = 4.19DD178 pKa = 4.83KK179 pKa = 11.76KK180 pKa = 11.17DD181 pKa = 3.38SDD183 pKa = 4.28VNEE186 pKa = 4.29TPPLDD191 pKa = 3.77TEE193 pKa = 4.6EE194 pKa = 4.59NDD196 pKa = 3.54LNKK199 pKa = 10.59GKK201 pKa = 10.11EE202 pKa = 4.28GPEE205 pKa = 3.59IRR207 pKa = 11.84DD208 pKa = 3.48DD209 pKa = 3.57KK210 pKa = 11.16SQNEE214 pKa = 4.74GIGDD218 pKa = 3.88VNSVVQKK225 pKa = 9.44TLEE228 pKa = 4.37PQSSTEE234 pKa = 3.98EE235 pKa = 4.14NGVEE239 pKa = 4.32TNDD242 pKa = 3.74DD243 pKa = 3.72TSVDD247 pKa = 3.55PVTNPEE253 pKa = 3.95MKK255 pKa = 10.63NEE257 pKa = 4.04VGEE260 pKa = 4.23ATHH263 pKa = 6.47QDD265 pKa = 2.89VSMEE269 pKa = 4.09AKK271 pKa = 10.37GDD273 pKa = 3.76LGKK276 pKa = 11.13GPNFDD281 pKa = 3.6TSKK284 pKa = 10.96EE285 pKa = 4.24SGDD288 pKa = 3.89SKK290 pKa = 11.55GLDD293 pKa = 3.63GGNDD297 pKa = 3.69DD298 pKa = 5.51QISDD302 pKa = 4.23GNSDD306 pKa = 3.78RR307 pKa = 11.84YY308 pKa = 10.56SEE310 pKa = 4.14EE311 pKa = 3.8AIKK314 pKa = 10.57RR315 pKa = 11.84LEE317 pKa = 3.95QMQGEE322 pKa = 4.15LSEE325 pKa = 4.46FEE327 pKa = 5.04LRR329 pKa = 11.84LKK331 pKa = 10.63QLIDD335 pKa = 3.22
MM1 pKa = 7.4LPVATVTTATILTTSLISSGKK22 pKa = 9.89VDD24 pKa = 4.52LDD26 pKa = 3.44KK27 pKa = 11.4KK28 pKa = 10.95KK29 pKa = 10.83EE30 pKa = 4.04NEE32 pKa = 4.01DD33 pKa = 3.31QEE35 pKa = 5.8LNDD38 pKa = 3.84FVKK41 pKa = 10.63PPEE44 pKa = 4.34DD45 pKa = 4.09FEE47 pKa = 4.77DD48 pKa = 3.56EE49 pKa = 4.36KK50 pKa = 11.08IFYY53 pKa = 10.5SKK55 pKa = 11.03EE56 pKa = 3.75GDD58 pKa = 3.48SDD60 pKa = 3.84NLRR63 pKa = 11.84TLDD66 pKa = 3.83KK67 pKa = 10.54DD68 pKa = 3.6TAKK71 pKa = 10.7KK72 pKa = 10.15IEE74 pKa = 4.17EE75 pKa = 4.05LMQKK79 pKa = 10.38NGGKK83 pKa = 9.76LEE85 pKa = 4.28KK86 pKa = 10.8GEE88 pKa = 4.03QLGDD92 pKa = 3.46TVQAPNPEE100 pKa = 4.11INPNVPEE107 pKa = 4.1NQPQNVGKK115 pKa = 10.26ADD117 pKa = 3.68SPEE120 pKa = 3.97ITSKK124 pKa = 11.48AEE126 pKa = 4.24DD127 pKa = 3.61KK128 pKa = 11.49DD129 pKa = 3.67MTSVPKK135 pKa = 10.82GNNKK139 pKa = 10.3DD140 pKa = 3.5EE141 pKa = 4.83GDD143 pKa = 4.1TISEE147 pKa = 4.4TKK149 pKa = 9.54DD150 pKa = 3.17TLISTSEE157 pKa = 4.02STDD160 pKa = 3.51DD161 pKa = 4.67DD162 pKa = 4.08FTNGRR167 pKa = 11.84DD168 pKa = 4.0DD169 pKa = 4.01PQQPSLDD176 pKa = 4.04DD177 pKa = 4.19DD178 pKa = 4.83KK179 pKa = 11.76KK180 pKa = 11.17DD181 pKa = 3.38SDD183 pKa = 4.28VNEE186 pKa = 4.29TPPLDD191 pKa = 3.77TEE193 pKa = 4.6EE194 pKa = 4.59NDD196 pKa = 3.54LNKK199 pKa = 10.59GKK201 pKa = 10.11EE202 pKa = 4.28GPEE205 pKa = 3.59IRR207 pKa = 11.84DD208 pKa = 3.48DD209 pKa = 3.57KK210 pKa = 11.16SQNEE214 pKa = 4.74GIGDD218 pKa = 3.88VNSVVQKK225 pKa = 9.44TLEE228 pKa = 4.37PQSSTEE234 pKa = 3.98EE235 pKa = 4.14NGVEE239 pKa = 4.32TNDD242 pKa = 3.74DD243 pKa = 3.72TSVDD247 pKa = 3.55PVTNPEE253 pKa = 3.95MKK255 pKa = 10.63NEE257 pKa = 4.04VGEE260 pKa = 4.23ATHH263 pKa = 6.47QDD265 pKa = 2.89VSMEE269 pKa = 4.09AKK271 pKa = 10.37GDD273 pKa = 3.76LGKK276 pKa = 11.13GPNFDD281 pKa = 3.6TSKK284 pKa = 10.96EE285 pKa = 4.24SGDD288 pKa = 3.89SKK290 pKa = 11.55GLDD293 pKa = 3.63GGNDD297 pKa = 3.69DD298 pKa = 5.51QISDD302 pKa = 4.23GNSDD306 pKa = 3.78RR307 pKa = 11.84YY308 pKa = 10.56SEE310 pKa = 4.14EE311 pKa = 3.8AIKK314 pKa = 10.57RR315 pKa = 11.84LEE317 pKa = 3.95QMQGEE322 pKa = 4.15LSEE325 pKa = 4.46FEE327 pKa = 5.04LRR329 pKa = 11.84LKK331 pKa = 10.63QLIDD335 pKa = 3.22
Molecular weight: 36.73 kDa
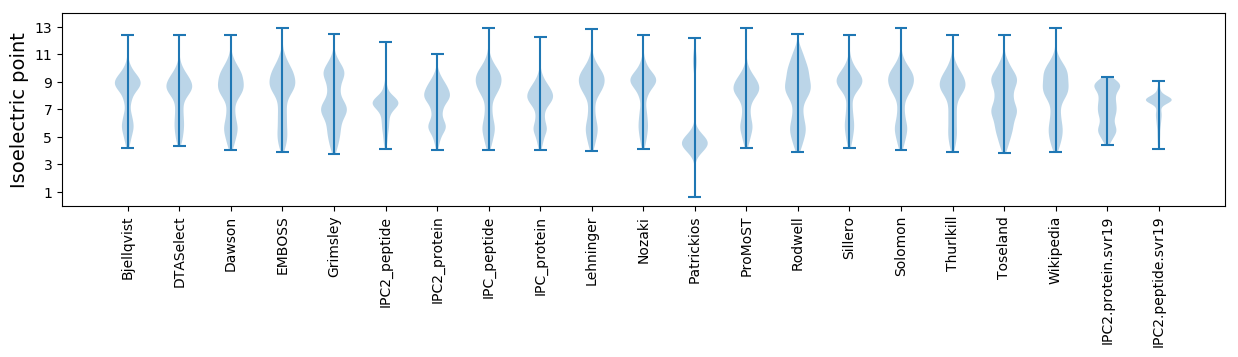
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A9QES2|A0A1A9QES2_9MOLU Uncharacterized protein OS=Candidatus Mycoplasma haemobos OX=432608 GN=A6V39_01990 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.3IKK4 pKa = 10.44IKK6 pKa = 9.21TKK8 pKa = 9.45KK9 pKa = 10.26AFSKK13 pKa = 10.58RR14 pKa = 11.84IRR16 pKa = 11.84VLGNGKK22 pKa = 9.75LKK24 pKa = 10.38RR25 pKa = 11.84KK26 pKa = 9.23HH27 pKa = 4.81SHH29 pKa = 6.44RR30 pKa = 11.84SHH32 pKa = 6.72LASNKK37 pKa = 7.0STKK40 pKa = 8.29QKK42 pKa = 9.73RR43 pKa = 11.84QARR46 pKa = 11.84SSVMLNKK53 pKa = 10.26SQIKK57 pKa = 9.73RR58 pKa = 11.84AYY60 pKa = 9.94QLLQGG65 pKa = 4.28
MM1 pKa = 7.62KK2 pKa = 10.3IKK4 pKa = 10.44IKK6 pKa = 9.21TKK8 pKa = 9.45KK9 pKa = 10.26AFSKK13 pKa = 10.58RR14 pKa = 11.84IRR16 pKa = 11.84VLGNGKK22 pKa = 9.75LKK24 pKa = 10.38RR25 pKa = 11.84KK26 pKa = 9.23HH27 pKa = 4.81SHH29 pKa = 6.44RR30 pKa = 11.84SHH32 pKa = 6.72LASNKK37 pKa = 7.0STKK40 pKa = 8.29QKK42 pKa = 9.73RR43 pKa = 11.84QARR46 pKa = 11.84SSVMLNKK53 pKa = 10.26SQIKK57 pKa = 9.73RR58 pKa = 11.84AYY60 pKa = 9.94QLLQGG65 pKa = 4.28
Molecular weight: 7.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
274467 |
55 |
2048 |
247.5 |
28.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.599 ± 0.067 | 1.317 ± 0.028 |
5.648 ± 0.064 | 7.725 ± 0.082 |
4.571 ± 0.079 | 5.581 ± 0.068 |
1.354 ± 0.027 | 7.313 ± 0.078 |
10.93 ± 0.1 | 9.529 ± 0.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.393 ± 0.027 | 6.424 ± 0.063 |
2.825 ± 0.044 | 3.136 ± 0.043 |
3.174 ± 0.047 | 7.428 ± 0.07 |
5.801 ± 0.075 | 5.011 ± 0.054 |
1.624 ± 0.035 | 3.615 ± 0.056 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
