
Dorea sp. AF36-15AT
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Dorea; unclassified Dorea
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
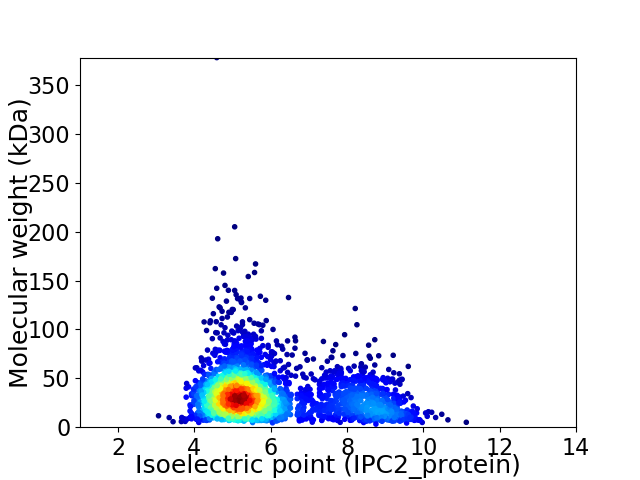
Virtual 2D-PAGE plot for 2785 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A396L2J9|A0A396L2J9_9FIRM Histidine kinase OS=Dorea sp. AF36-15AT OX=2292041 GN=DWZ93_06190 PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 10.64NRR4 pKa = 11.84ITRR7 pKa = 11.84IAMTLTMLMVPALVTGCGPSKK28 pKa = 10.65KK29 pKa = 10.0QIQQEE34 pKa = 4.01LSYY37 pKa = 10.94QEE39 pKa = 4.28PDD41 pKa = 3.09QEE43 pKa = 4.34EE44 pKa = 4.1LHH46 pKa = 7.0EE47 pKa = 4.69EE48 pKa = 4.05YY49 pKa = 10.64QYY51 pKa = 11.32LAEE54 pKa = 4.22VNEE57 pKa = 4.42GDD59 pKa = 4.07GVATGLINEE68 pKa = 4.33YY69 pKa = 10.08GEE71 pKa = 4.72CVVACMFDD79 pKa = 4.42EE80 pKa = 4.33ITMGMTDD87 pKa = 3.52DD88 pKa = 4.19GFRR91 pKa = 11.84ILAYY95 pKa = 9.89KK96 pKa = 10.01SVYY99 pKa = 8.65TDD101 pKa = 3.16KK102 pKa = 11.15QGEE105 pKa = 4.43SEE107 pKa = 4.14AYY109 pKa = 9.28RR110 pKa = 11.84YY111 pKa = 9.86YY112 pKa = 11.27VLDD115 pKa = 3.34EE116 pKa = 4.52RR117 pKa = 11.84GQILEE122 pKa = 3.96EE123 pKa = 4.75HH124 pKa = 6.66GEE126 pKa = 4.39GIEE129 pKa = 4.09EE130 pKa = 4.16EE131 pKa = 4.3GGGYY135 pKa = 10.14YY136 pKa = 10.46DD137 pKa = 5.37GEE139 pKa = 4.39ALLEE143 pKa = 4.28EE144 pKa = 4.61MDD146 pKa = 3.79NYY148 pKa = 10.83TPVEE152 pKa = 4.16VEE154 pKa = 4.02DD155 pKa = 4.1AVSEE159 pKa = 4.12EE160 pKa = 4.39AEE162 pKa = 4.28EE163 pKa = 4.13EE164 pKa = 4.06TAEE167 pKa = 4.36ADD169 pKa = 3.66SAPTDD174 pKa = 3.72VEE176 pKa = 4.71IVTTDD181 pKa = 3.25DD182 pKa = 3.28GDD184 pKa = 4.11YY185 pKa = 11.36EE186 pKa = 5.36LVDD189 pKa = 3.32QSGKK193 pKa = 10.21VLISDD198 pKa = 3.82EE199 pKa = 4.61DD200 pKa = 4.02VSGMGVSDD208 pKa = 4.87IISMYY213 pKa = 8.23FTRR216 pKa = 11.84DD217 pKa = 2.97GKK219 pKa = 10.74YY220 pKa = 10.47VIADD224 pKa = 4.03TGSMLYY230 pKa = 10.59DD231 pKa = 3.89EE232 pKa = 6.07DD233 pKa = 4.16EE234 pKa = 4.55LPIGGNVLFDD244 pKa = 3.91LEE246 pKa = 4.87GNVKK250 pKa = 10.32RR251 pKa = 11.84SNDD254 pKa = 3.68YY255 pKa = 11.09EE256 pKa = 4.59SISYY260 pKa = 10.77GGDD263 pKa = 3.3NGWLCVHH270 pKa = 6.43NKK272 pKa = 8.08KK273 pKa = 8.77TNQTVYY279 pKa = 10.9LNSDD283 pKa = 4.17LKK285 pKa = 10.51TEE287 pKa = 4.59LDD289 pKa = 3.56LGDD292 pKa = 4.34AYY294 pKa = 10.66GAAGDD299 pKa = 3.89FKK301 pKa = 11.19KK302 pKa = 10.66CLRR305 pKa = 11.84RR306 pKa = 11.84QQ307 pKa = 3.21
MM1 pKa = 7.76KK2 pKa = 10.64NRR4 pKa = 11.84ITRR7 pKa = 11.84IAMTLTMLMVPALVTGCGPSKK28 pKa = 10.65KK29 pKa = 10.0QIQQEE34 pKa = 4.01LSYY37 pKa = 10.94QEE39 pKa = 4.28PDD41 pKa = 3.09QEE43 pKa = 4.34EE44 pKa = 4.1LHH46 pKa = 7.0EE47 pKa = 4.69EE48 pKa = 4.05YY49 pKa = 10.64QYY51 pKa = 11.32LAEE54 pKa = 4.22VNEE57 pKa = 4.42GDD59 pKa = 4.07GVATGLINEE68 pKa = 4.33YY69 pKa = 10.08GEE71 pKa = 4.72CVVACMFDD79 pKa = 4.42EE80 pKa = 4.33ITMGMTDD87 pKa = 3.52DD88 pKa = 4.19GFRR91 pKa = 11.84ILAYY95 pKa = 9.89KK96 pKa = 10.01SVYY99 pKa = 8.65TDD101 pKa = 3.16KK102 pKa = 11.15QGEE105 pKa = 4.43SEE107 pKa = 4.14AYY109 pKa = 9.28RR110 pKa = 11.84YY111 pKa = 9.86YY112 pKa = 11.27VLDD115 pKa = 3.34EE116 pKa = 4.52RR117 pKa = 11.84GQILEE122 pKa = 3.96EE123 pKa = 4.75HH124 pKa = 6.66GEE126 pKa = 4.39GIEE129 pKa = 4.09EE130 pKa = 4.16EE131 pKa = 4.3GGGYY135 pKa = 10.14YY136 pKa = 10.46DD137 pKa = 5.37GEE139 pKa = 4.39ALLEE143 pKa = 4.28EE144 pKa = 4.61MDD146 pKa = 3.79NYY148 pKa = 10.83TPVEE152 pKa = 4.16VEE154 pKa = 4.02DD155 pKa = 4.1AVSEE159 pKa = 4.12EE160 pKa = 4.39AEE162 pKa = 4.28EE163 pKa = 4.13EE164 pKa = 4.06TAEE167 pKa = 4.36ADD169 pKa = 3.66SAPTDD174 pKa = 3.72VEE176 pKa = 4.71IVTTDD181 pKa = 3.25DD182 pKa = 3.28GDD184 pKa = 4.11YY185 pKa = 11.36EE186 pKa = 5.36LVDD189 pKa = 3.32QSGKK193 pKa = 10.21VLISDD198 pKa = 3.82EE199 pKa = 4.61DD200 pKa = 4.02VSGMGVSDD208 pKa = 4.87IISMYY213 pKa = 8.23FTRR216 pKa = 11.84DD217 pKa = 2.97GKK219 pKa = 10.74YY220 pKa = 10.47VIADD224 pKa = 4.03TGSMLYY230 pKa = 10.59DD231 pKa = 3.89EE232 pKa = 6.07DD233 pKa = 4.16EE234 pKa = 4.55LPIGGNVLFDD244 pKa = 3.91LEE246 pKa = 4.87GNVKK250 pKa = 10.32RR251 pKa = 11.84SNDD254 pKa = 3.68YY255 pKa = 11.09EE256 pKa = 4.59SISYY260 pKa = 10.77GGDD263 pKa = 3.3NGWLCVHH270 pKa = 6.43NKK272 pKa = 8.08KK273 pKa = 8.77TNQTVYY279 pKa = 10.9LNSDD283 pKa = 4.17LKK285 pKa = 10.51TEE287 pKa = 4.59LDD289 pKa = 3.56LGDD292 pKa = 4.34AYY294 pKa = 10.66GAAGDD299 pKa = 3.89FKK301 pKa = 11.19KK302 pKa = 10.66CLRR305 pKa = 11.84RR306 pKa = 11.84QQ307 pKa = 3.21
Molecular weight: 34.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A396L5D0|A0A396L5D0_9FIRM ImmA/IrrE family metallo-endopeptidase OS=Dorea sp. AF36-15AT OX=2292041 GN=DWZ93_11500 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.87VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 8.85KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.87VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 8.85KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
871764 |
27 |
3528 |
313.0 |
35.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.565 ± 0.05 | 1.533 ± 0.021 |
5.833 ± 0.037 | 7.849 ± 0.059 |
3.751 ± 0.036 | 7.218 ± 0.041 |
1.856 ± 0.02 | 7.31 ± 0.044 |
6.877 ± 0.046 | 8.694 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.306 ± 0.026 | 4.154 ± 0.031 |
3.262 ± 0.027 | 3.295 ± 0.028 |
4.47 ± 0.044 | 5.473 ± 0.032 |
5.469 ± 0.048 | 7.122 ± 0.042 |
0.861 ± 0.018 | 4.103 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
