
Ajellomyces capsulatus (strain H143) (Darling s disease fungus) (Histoplasma capsulatum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Onygenales; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
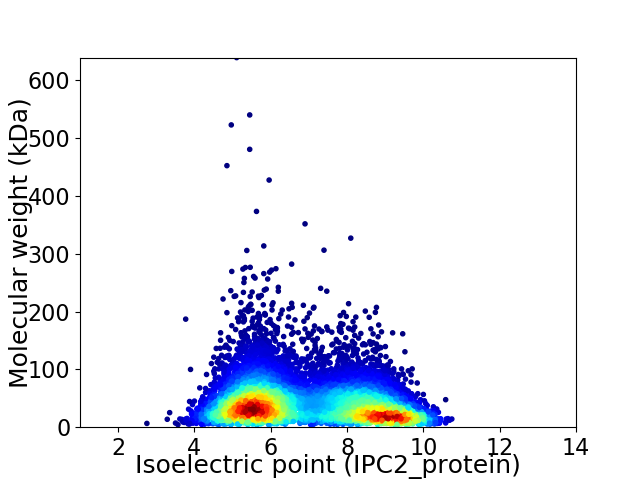
Virtual 2D-PAGE plot for 9545 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6HAB4|C6HAB4_AJECH RNA binding protein OS=Ajellomyces capsulatus (strain H143) OX=544712 GN=HCDG_03145 PE=4 SV=1
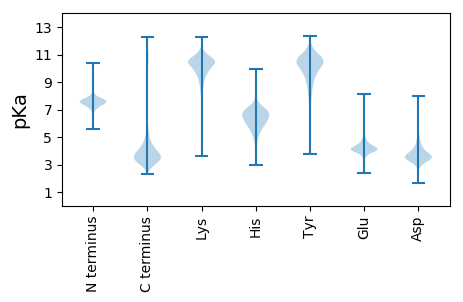
MM1 pKa = 6.88QWVLHH6 pKa = 6.39RR7 pKa = 11.84LTALSGGADD16 pKa = 3.59VLEE19 pKa = 5.23LDD21 pKa = 5.52LAPDD25 pKa = 4.8DD26 pKa = 4.52NDD28 pKa = 4.12SDD30 pKa = 4.25GSSHH34 pKa = 7.14LNISDD39 pKa = 3.73EE40 pKa = 4.47TPLTEE45 pKa = 4.42GVTLKK50 pKa = 10.63EE51 pKa = 3.89YY52 pKa = 10.92LDD54 pKa = 3.66TYY56 pKa = 10.73EE57 pKa = 4.43YY58 pKa = 11.23GSTEE62 pKa = 4.63INDD65 pKa = 2.79GWYY68 pKa = 9.64RR69 pKa = 11.84SQIIGSS75 pKa = 3.74
MM1 pKa = 6.88QWVLHH6 pKa = 6.39RR7 pKa = 11.84LTALSGGADD16 pKa = 3.59VLEE19 pKa = 5.23LDD21 pKa = 5.52LAPDD25 pKa = 4.8DD26 pKa = 4.52NDD28 pKa = 4.12SDD30 pKa = 4.25GSSHH34 pKa = 7.14LNISDD39 pKa = 3.73EE40 pKa = 4.47TPLTEE45 pKa = 4.42GVTLKK50 pKa = 10.63EE51 pKa = 3.89YY52 pKa = 10.92LDD54 pKa = 3.66TYY56 pKa = 10.73EE57 pKa = 4.43YY58 pKa = 11.23GSTEE62 pKa = 4.63INDD65 pKa = 2.79GWYY68 pKa = 9.64RR69 pKa = 11.84SQIIGSS75 pKa = 3.74
Molecular weight: 8.29 kDa
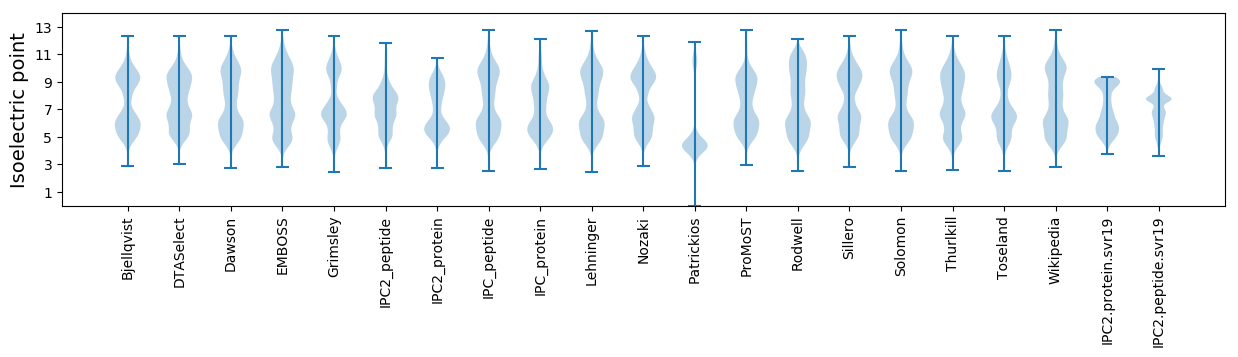
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6HK97|C6HK97_AJECH Uncharacterized protein OS=Ajellomyces capsulatus (strain H143) OX=544712 GN=HCDG_06628 PE=4 SV=1
MM1 pKa = 7.93KK2 pKa = 10.07SQQWTPSIHH11 pKa = 5.29STTRR15 pKa = 11.84SLANRR20 pKa = 11.84HH21 pKa = 5.9SGAILQEE28 pKa = 4.41SGPVPLPPRR37 pKa = 11.84LYY39 pKa = 11.0GPGSLPTTGRR49 pKa = 11.84SQGTYY54 pKa = 10.14DD55 pKa = 3.39PLGRR59 pKa = 11.84RR60 pKa = 11.84EE61 pKa = 4.29NEE63 pKa = 4.0LSPAAPSGPPPSSLIYY79 pKa = 9.79PPQAYY84 pKa = 8.17HH85 pKa = 7.21HH86 pKa = 6.62EE87 pKa = 4.79IIRR90 pKa = 11.84HH91 pKa = 5.8AKK93 pKa = 9.21PSPPPGPFASMHH105 pKa = 6.46PEE107 pKa = 3.87PTPSFHH113 pKa = 7.02SRR115 pKa = 11.84HH116 pKa = 5.6SSRR119 pKa = 11.84GSVIKK124 pKa = 10.64GGEE127 pKa = 4.29GVKK130 pKa = 10.64EE131 pKa = 4.14PGNALYY137 pKa = 10.72RR138 pKa = 11.84EE139 pKa = 4.25DD140 pKa = 3.6TNIFVGVFLEE150 pKa = 4.3PWYY153 pKa = 10.77SGRR156 pKa = 11.84DD157 pKa = 3.46SNKK160 pKa = 8.3PAKK163 pKa = 10.09LRR165 pKa = 11.84ACIEE169 pKa = 4.16TLLCLSPRR177 pKa = 11.84PTHH180 pKa = 6.81HH181 pKa = 6.44EE182 pKa = 3.74QTEE185 pKa = 4.09PSSNRR190 pKa = 11.84LATKK194 pKa = 10.03QAAKK198 pKa = 10.23FPSPQVSLTQPPHH211 pKa = 5.7VKK213 pKa = 10.2LIHH216 pKa = 6.08HH217 pKa = 6.74HH218 pKa = 5.66LHH220 pKa = 6.67RR221 pKa = 11.84PPCRR225 pKa = 11.84RR226 pKa = 11.84GTCRR230 pKa = 11.84IPRR233 pKa = 11.84SPTSRR238 pKa = 11.84QGDD241 pKa = 4.25GLWAAMTVLLSILPQGKK258 pKa = 8.24LTTPAFSNRR267 pKa = 11.84LHH269 pKa = 7.11PLRR272 pKa = 11.84PTIVARR278 pKa = 11.84CLLHH282 pKa = 6.77LLPSTTTRR290 pKa = 11.84VEE292 pKa = 4.48APFQPPLLPNRR303 pKa = 11.84PPSLHH308 pKa = 6.36VNPPNHH314 pKa = 6.52PHH316 pKa = 5.62TARR319 pKa = 11.84VAACRR324 pKa = 11.84YY325 pKa = 6.99RR326 pKa = 11.84QCC328 pKa = 4.73
MM1 pKa = 7.93KK2 pKa = 10.07SQQWTPSIHH11 pKa = 5.29STTRR15 pKa = 11.84SLANRR20 pKa = 11.84HH21 pKa = 5.9SGAILQEE28 pKa = 4.41SGPVPLPPRR37 pKa = 11.84LYY39 pKa = 11.0GPGSLPTTGRR49 pKa = 11.84SQGTYY54 pKa = 10.14DD55 pKa = 3.39PLGRR59 pKa = 11.84RR60 pKa = 11.84EE61 pKa = 4.29NEE63 pKa = 4.0LSPAAPSGPPPSSLIYY79 pKa = 9.79PPQAYY84 pKa = 8.17HH85 pKa = 7.21HH86 pKa = 6.62EE87 pKa = 4.79IIRR90 pKa = 11.84HH91 pKa = 5.8AKK93 pKa = 9.21PSPPPGPFASMHH105 pKa = 6.46PEE107 pKa = 3.87PTPSFHH113 pKa = 7.02SRR115 pKa = 11.84HH116 pKa = 5.6SSRR119 pKa = 11.84GSVIKK124 pKa = 10.64GGEE127 pKa = 4.29GVKK130 pKa = 10.64EE131 pKa = 4.14PGNALYY137 pKa = 10.72RR138 pKa = 11.84EE139 pKa = 4.25DD140 pKa = 3.6TNIFVGVFLEE150 pKa = 4.3PWYY153 pKa = 10.77SGRR156 pKa = 11.84DD157 pKa = 3.46SNKK160 pKa = 8.3PAKK163 pKa = 10.09LRR165 pKa = 11.84ACIEE169 pKa = 4.16TLLCLSPRR177 pKa = 11.84PTHH180 pKa = 6.81HH181 pKa = 6.44EE182 pKa = 3.74QTEE185 pKa = 4.09PSSNRR190 pKa = 11.84LATKK194 pKa = 10.03QAAKK198 pKa = 10.23FPSPQVSLTQPPHH211 pKa = 5.7VKK213 pKa = 10.2LIHH216 pKa = 6.08HH217 pKa = 6.74HH218 pKa = 5.66LHH220 pKa = 6.67RR221 pKa = 11.84PPCRR225 pKa = 11.84RR226 pKa = 11.84GTCRR230 pKa = 11.84IPRR233 pKa = 11.84SPTSRR238 pKa = 11.84QGDD241 pKa = 4.25GLWAAMTVLLSILPQGKK258 pKa = 8.24LTTPAFSNRR267 pKa = 11.84LHH269 pKa = 7.11PLRR272 pKa = 11.84PTIVARR278 pKa = 11.84CLLHH282 pKa = 6.77LLPSTTTRR290 pKa = 11.84VEE292 pKa = 4.48APFQPPLLPNRR303 pKa = 11.84PPSLHH308 pKa = 6.36VNPPNHH314 pKa = 6.52PHH316 pKa = 5.62TARR319 pKa = 11.84VAACRR324 pKa = 11.84YY325 pKa = 6.99RR326 pKa = 11.84QCC328 pKa = 4.73
Molecular weight: 36.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3861477 |
39 |
5826 |
404.6 |
44.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.909 ± 0.021 | 1.288 ± 0.009 |
5.542 ± 0.018 | 6.135 ± 0.025 |
3.649 ± 0.017 | 6.716 ± 0.025 |
2.511 ± 0.011 | 5.119 ± 0.02 |
5.011 ± 0.021 | 8.897 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.107 ± 0.01 | 3.953 ± 0.015 |
6.236 ± 0.027 | 4.048 ± 0.019 |
6.463 ± 0.022 | 8.829 ± 0.033 |
5.852 ± 0.017 | 5.784 ± 0.017 |
1.335 ± 0.009 | 2.617 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
