
Human parvovirus B19 (strain HV) (HPV B19)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Parvovirinae; Erythroparvovirus; Primate erythroparvovirus 1; Human parvovirus B19
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
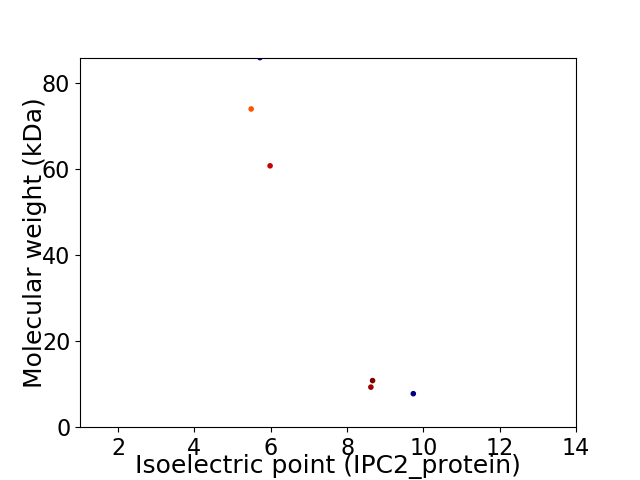
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9PZT0-2|CAPSD-2_PAVHV Isoform of Q9PZT0 Isoform Major capsid protein VP2 of Minor capsid protein VP1 OS=Human parvovirus B19 (strain HV) OX=648237 GN=vp PE=1 SV=1
MM1 pKa = 7.42EE2 pKa = 5.69LFRR5 pKa = 11.84GVLQVSSNVLDD16 pKa = 4.09CANDD20 pKa = 3.54NWWCSLLDD28 pKa = 4.52LDD30 pKa = 4.58TSDD33 pKa = 4.51WEE35 pKa = 4.51PLTHH39 pKa = 6.33TNRR42 pKa = 11.84LMAIYY47 pKa = 9.89LSSVASKK54 pKa = 11.22LDD56 pKa = 3.69FTGGPLAGCLYY67 pKa = 10.0FFQVEE72 pKa = 4.38CNKK75 pKa = 10.02FEE77 pKa = 4.32EE78 pKa = 6.0GYY80 pKa = 10.13HH81 pKa = 4.95IHH83 pKa = 6.23VVIGGPGLNPRR94 pKa = 11.84NLTVCVEE101 pKa = 4.15GLFNNVLYY109 pKa = 10.84HH110 pKa = 7.15LVTEE114 pKa = 4.21NVKK117 pKa = 10.88LKK119 pKa = 9.87FLPGMTTKK127 pKa = 10.67GKK129 pKa = 9.46YY130 pKa = 9.84FRR132 pKa = 11.84DD133 pKa = 3.76GEE135 pKa = 4.32QFIEE139 pKa = 4.17NYY141 pKa = 9.96LMKK144 pKa = 10.45KK145 pKa = 9.83IPLNVVWCVTNIDD158 pKa = 4.51GYY160 pKa = 10.72IDD162 pKa = 3.5TCISATFRR170 pKa = 11.84RR171 pKa = 11.84GACHH175 pKa = 6.39AKK177 pKa = 10.11KK178 pKa = 10.17PRR180 pKa = 11.84ITTAINDD187 pKa = 3.84TSSDD191 pKa = 3.41AGEE194 pKa = 4.2SSGTGAEE201 pKa = 3.92VVPINGKK208 pKa = 6.92GTKK211 pKa = 10.13ASIKK215 pKa = 9.62FQTMVNWLCEE225 pKa = 3.97NRR227 pKa = 11.84VFTEE231 pKa = 4.36DD232 pKa = 2.61KK233 pKa = 10.24WKK235 pKa = 10.84LVDD238 pKa = 4.25FNQYY242 pKa = 8.33TLLSSSHH249 pKa = 6.29SGSFQIQSALKK260 pKa = 9.41LAIYY264 pKa = 10.0KK265 pKa = 8.2ATNLVPTSTFLLHH278 pKa = 6.7TDD280 pKa = 4.73FEE282 pKa = 4.74QVMCIKK288 pKa = 10.5DD289 pKa = 3.64NKK291 pKa = 9.48IVKK294 pKa = 10.06LLLCQNYY301 pKa = 10.3DD302 pKa = 3.57PLLVGQHH309 pKa = 4.24VLKK312 pKa = 10.54WIDD315 pKa = 3.44KK316 pKa = 10.25KK317 pKa = 11.02CGKK320 pKa = 10.44KK321 pKa = 8.43NTLWFYY327 pKa = 10.95GPPSTGKK334 pKa = 8.67TNLAMAIAKK343 pKa = 8.26SVPVYY348 pKa = 11.35GMVNWNNEE356 pKa = 3.9NFPFNDD362 pKa = 3.11VAGKK366 pKa = 10.34SLVVWDD372 pKa = 4.04EE373 pKa = 4.53GIIKK377 pKa = 8.56STIVEE382 pKa = 3.9AAKK385 pKa = 10.54AILGGQPTRR394 pKa = 11.84VDD396 pKa = 2.94QKK398 pKa = 9.77MRR400 pKa = 11.84GSVAVPGVPVVITSNGDD417 pKa = 2.82ITFVVSGNTTTTVHH431 pKa = 6.73AKK433 pKa = 9.85ALKK436 pKa = 10.0EE437 pKa = 3.91RR438 pKa = 11.84MVKK441 pKa = 10.64LNFTVRR447 pKa = 11.84CSPDD451 pKa = 2.98MGLLTEE457 pKa = 5.44ADD459 pKa = 3.75VQQWLTWCNAQSWDD473 pKa = 4.16HH474 pKa = 5.84YY475 pKa = 10.07EE476 pKa = 3.7NWAINYY482 pKa = 7.53TFDD485 pKa = 4.51FPGINADD492 pKa = 3.84ALHH495 pKa = 7.51PDD497 pKa = 3.8LQTTPIVTDD506 pKa = 3.41TSISSSGGEE515 pKa = 3.93SSEE518 pKa = 4.12EE519 pKa = 3.7LSEE522 pKa = 4.59SSFFNLITPGAWNTEE537 pKa = 4.41TPRR540 pKa = 11.84SSTPIPGTSSGEE552 pKa = 4.22SFVGSSVSSEE562 pKa = 3.84VVAASWEE569 pKa = 4.22EE570 pKa = 3.88AFYY573 pKa = 10.7TPLADD578 pKa = 3.63QFRR581 pKa = 11.84EE582 pKa = 4.0LLVGVDD588 pKa = 3.93YY589 pKa = 11.4VWDD592 pKa = 4.18GVRR595 pKa = 11.84GLPVCCVQHH604 pKa = 6.43INNSGGGLGLCPHH617 pKa = 7.82CINVGAWYY625 pKa = 10.11NGWKK629 pKa = 9.97FRR631 pKa = 11.84EE632 pKa = 4.2FTPDD636 pKa = 3.72LVRR639 pKa = 11.84CSCHH643 pKa = 6.95VGASNPFSVLTCKK656 pKa = 10.3KK657 pKa = 9.69CAYY660 pKa = 10.47LSGLQSFVDD669 pKa = 4.03YY670 pKa = 10.7EE671 pKa = 4.05
MM1 pKa = 7.42EE2 pKa = 5.69LFRR5 pKa = 11.84GVLQVSSNVLDD16 pKa = 4.09CANDD20 pKa = 3.54NWWCSLLDD28 pKa = 4.52LDD30 pKa = 4.58TSDD33 pKa = 4.51WEE35 pKa = 4.51PLTHH39 pKa = 6.33TNRR42 pKa = 11.84LMAIYY47 pKa = 9.89LSSVASKK54 pKa = 11.22LDD56 pKa = 3.69FTGGPLAGCLYY67 pKa = 10.0FFQVEE72 pKa = 4.38CNKK75 pKa = 10.02FEE77 pKa = 4.32EE78 pKa = 6.0GYY80 pKa = 10.13HH81 pKa = 4.95IHH83 pKa = 6.23VVIGGPGLNPRR94 pKa = 11.84NLTVCVEE101 pKa = 4.15GLFNNVLYY109 pKa = 10.84HH110 pKa = 7.15LVTEE114 pKa = 4.21NVKK117 pKa = 10.88LKK119 pKa = 9.87FLPGMTTKK127 pKa = 10.67GKK129 pKa = 9.46YY130 pKa = 9.84FRR132 pKa = 11.84DD133 pKa = 3.76GEE135 pKa = 4.32QFIEE139 pKa = 4.17NYY141 pKa = 9.96LMKK144 pKa = 10.45KK145 pKa = 9.83IPLNVVWCVTNIDD158 pKa = 4.51GYY160 pKa = 10.72IDD162 pKa = 3.5TCISATFRR170 pKa = 11.84RR171 pKa = 11.84GACHH175 pKa = 6.39AKK177 pKa = 10.11KK178 pKa = 10.17PRR180 pKa = 11.84ITTAINDD187 pKa = 3.84TSSDD191 pKa = 3.41AGEE194 pKa = 4.2SSGTGAEE201 pKa = 3.92VVPINGKK208 pKa = 6.92GTKK211 pKa = 10.13ASIKK215 pKa = 9.62FQTMVNWLCEE225 pKa = 3.97NRR227 pKa = 11.84VFTEE231 pKa = 4.36DD232 pKa = 2.61KK233 pKa = 10.24WKK235 pKa = 10.84LVDD238 pKa = 4.25FNQYY242 pKa = 8.33TLLSSSHH249 pKa = 6.29SGSFQIQSALKK260 pKa = 9.41LAIYY264 pKa = 10.0KK265 pKa = 8.2ATNLVPTSTFLLHH278 pKa = 6.7TDD280 pKa = 4.73FEE282 pKa = 4.74QVMCIKK288 pKa = 10.5DD289 pKa = 3.64NKK291 pKa = 9.48IVKK294 pKa = 10.06LLLCQNYY301 pKa = 10.3DD302 pKa = 3.57PLLVGQHH309 pKa = 4.24VLKK312 pKa = 10.54WIDD315 pKa = 3.44KK316 pKa = 10.25KK317 pKa = 11.02CGKK320 pKa = 10.44KK321 pKa = 8.43NTLWFYY327 pKa = 10.95GPPSTGKK334 pKa = 8.67TNLAMAIAKK343 pKa = 8.26SVPVYY348 pKa = 11.35GMVNWNNEE356 pKa = 3.9NFPFNDD362 pKa = 3.11VAGKK366 pKa = 10.34SLVVWDD372 pKa = 4.04EE373 pKa = 4.53GIIKK377 pKa = 8.56STIVEE382 pKa = 3.9AAKK385 pKa = 10.54AILGGQPTRR394 pKa = 11.84VDD396 pKa = 2.94QKK398 pKa = 9.77MRR400 pKa = 11.84GSVAVPGVPVVITSNGDD417 pKa = 2.82ITFVVSGNTTTTVHH431 pKa = 6.73AKK433 pKa = 9.85ALKK436 pKa = 10.0EE437 pKa = 3.91RR438 pKa = 11.84MVKK441 pKa = 10.64LNFTVRR447 pKa = 11.84CSPDD451 pKa = 2.98MGLLTEE457 pKa = 5.44ADD459 pKa = 3.75VQQWLTWCNAQSWDD473 pKa = 4.16HH474 pKa = 5.84YY475 pKa = 10.07EE476 pKa = 3.7NWAINYY482 pKa = 7.53TFDD485 pKa = 4.51FPGINADD492 pKa = 3.84ALHH495 pKa = 7.51PDD497 pKa = 3.8LQTTPIVTDD506 pKa = 3.41TSISSSGGEE515 pKa = 3.93SSEE518 pKa = 4.12EE519 pKa = 3.7LSEE522 pKa = 4.59SSFFNLITPGAWNTEE537 pKa = 4.41TPRR540 pKa = 11.84SSTPIPGTSSGEE552 pKa = 4.22SFVGSSVSSEE562 pKa = 3.84VVAASWEE569 pKa = 4.22EE570 pKa = 3.88AFYY573 pKa = 10.7TPLADD578 pKa = 3.63QFRR581 pKa = 11.84EE582 pKa = 4.0LLVGVDD588 pKa = 3.93YY589 pKa = 11.4VWDD592 pKa = 4.18GVRR595 pKa = 11.84GLPVCCVQHH604 pKa = 6.43INNSGGGLGLCPHH617 pKa = 7.82CINVGAWYY625 pKa = 10.11NGWKK629 pKa = 9.97FRR631 pKa = 11.84EE632 pKa = 4.2FTPDD636 pKa = 3.72LVRR639 pKa = 11.84CSCHH643 pKa = 6.95VGASNPFSVLTCKK656 pKa = 10.3KK657 pKa = 9.69CAYY660 pKa = 10.47LSGLQSFVDD669 pKa = 4.03YY670 pKa = 10.7EE671 pKa = 4.05
Molecular weight: 74.07 kDa
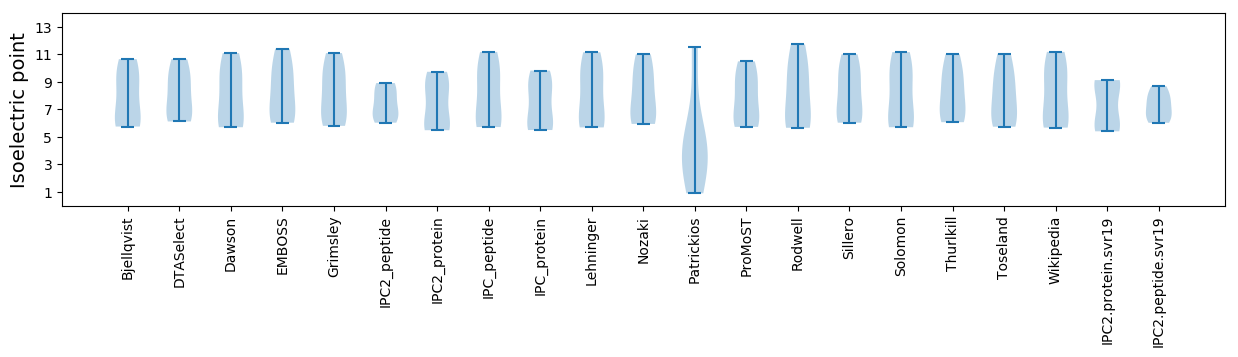
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0DJZ0|11K_PAVHV Host-modulation protein 11K OS=Human parvovirus B19 (strain HV) OX=648237 GN=11K PE=1 SV=1
MM1 pKa = 7.29QMPSTQTSKK10 pKa = 10.77PPQLSQTPVSAAVVVKK26 pKa = 10.53ALKK29 pKa = 10.54NSVKK33 pKa = 10.5AAFLTSSPQAPGTLKK48 pKa = 10.27PRR50 pKa = 11.84ALVRR54 pKa = 11.84PSPGPVQEE62 pKa = 4.2NHH64 pKa = 6.39LSEE67 pKa = 4.77AQFPPKK73 pKa = 10.4LL74 pKa = 3.64
MM1 pKa = 7.29QMPSTQTSKK10 pKa = 10.77PPQLSQTPVSAAVVVKK26 pKa = 10.53ALKK29 pKa = 10.54NSVKK33 pKa = 10.5AAFLTSSPQAPGTLKK48 pKa = 10.27PRR50 pKa = 11.84ALVRR54 pKa = 11.84PSPGPVQEE62 pKa = 4.2NHH64 pKa = 6.39LSEE67 pKa = 4.77AQFPPKK73 pKa = 10.4LL74 pKa = 3.64
Molecular weight: 7.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2255 |
74 |
781 |
375.8 |
41.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.987 ± 0.579 | 1.596 ± 0.718 |
4.169 ± 0.61 | 4.745 ± 0.364 |
3.858 ± 0.54 | 8.115 ± 0.961 |
3.193 ± 0.522 | 3.769 ± 0.56 |
5.543 ± 0.477 | 8.603 ± 0.756 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.173 ± 0.726 | 4.922 ± 0.511 |
7.361 ± 1.455 | 4.789 ± 0.76 |
2.661 ± 0.326 | 8.204 ± 0.729 |
7.273 ± 0.337 | 6.874 ± 0.775 |
1.863 ± 0.396 | 4.302 ± 0.707 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
